Investigating scaffold protein mutations and disease
There is a significant amount of research focused on diseases which are caused by mutations in scaffold proteins, like Daple, that regulate crosstalk between G protein signaling and other growth promoting pathways [1,2]. Mutations in these proteins have been linked to abnormalities like hydrocephalus and several cancers [1,2]. Many cancer researchers investigate protein signaling in model organisms like zebrafish using genetically modified fluorescent strains with the hopes of gaining a better mechanistic understanding of how faulty signaling causes disease [1,2].
Challenges when imaging model organisms
One of the limitations when imaging model organisms is the autofluorescence seen in some of the zebrafish tissues and the time it takes to image them. Conventionally, most of the imaging is done using confocal microscopy to achieve adequate resolution [1,2]. This involves acquisition of z-stacks in multiple fluorescent channels and would take on average 10-15 minutes to acquire a confocal image.
Methods
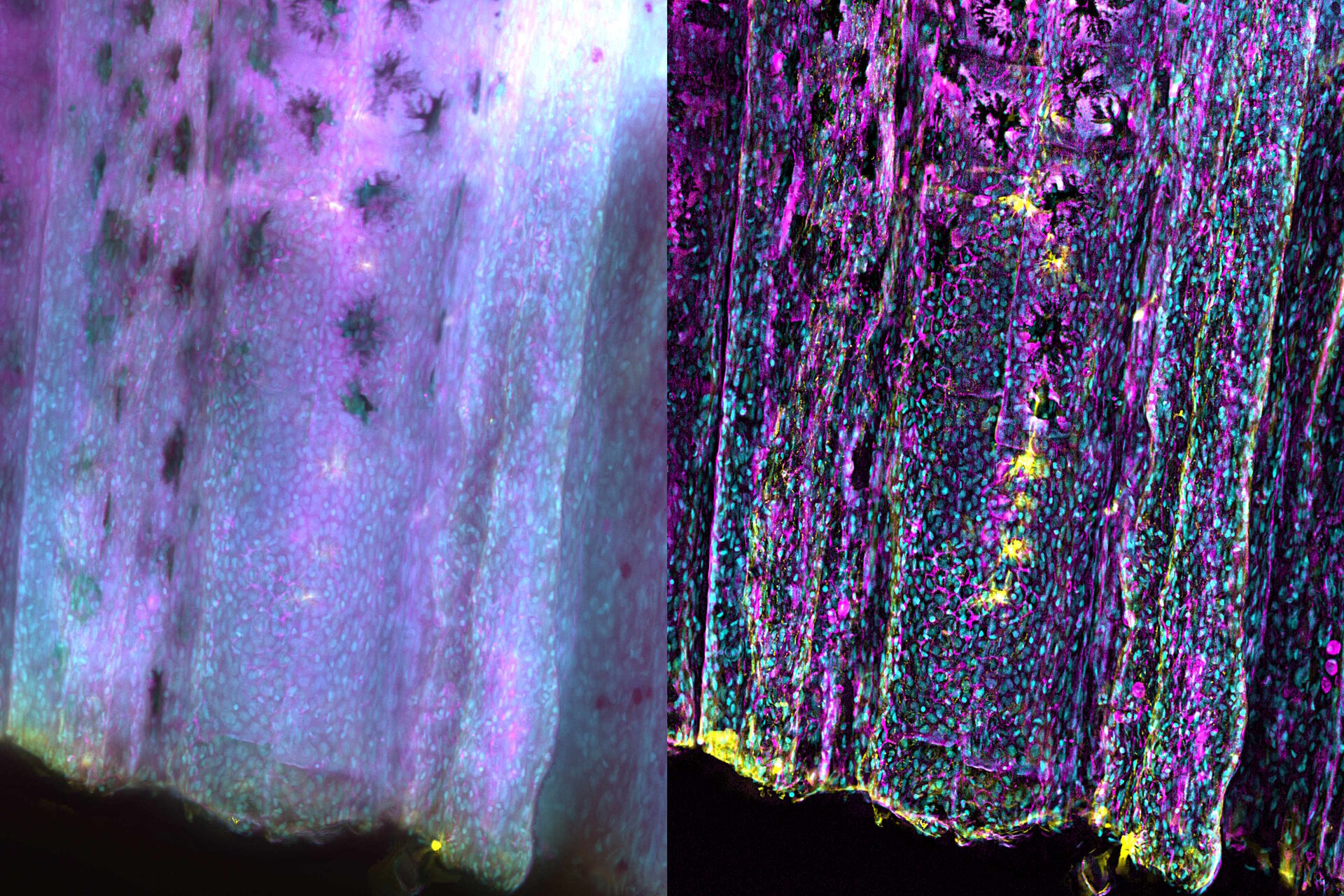
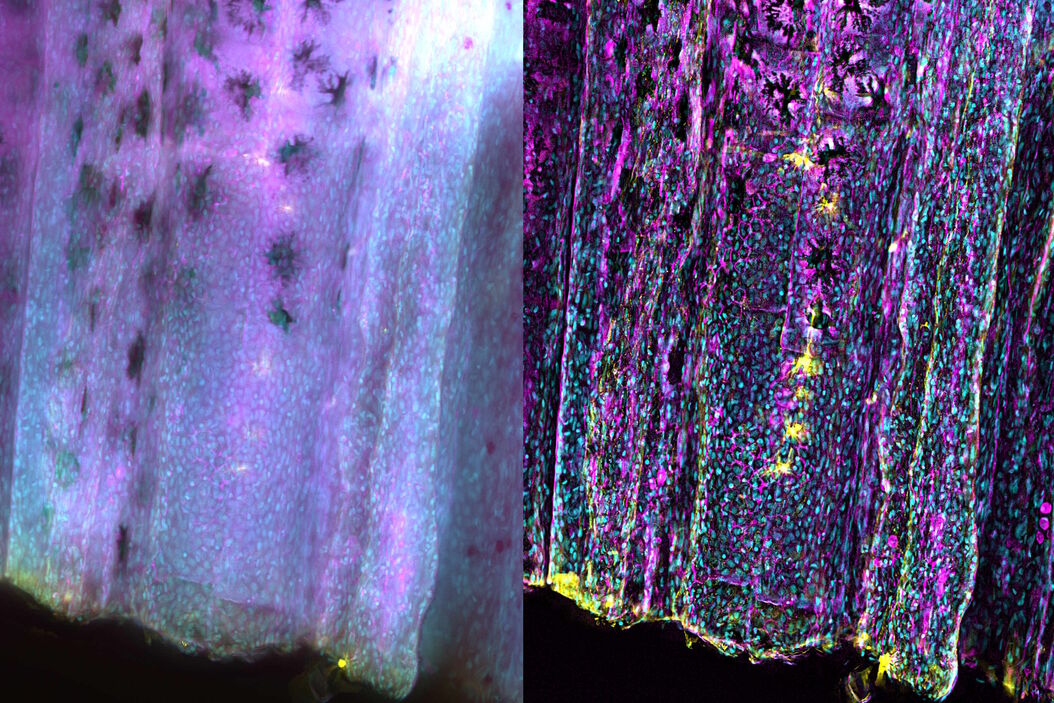
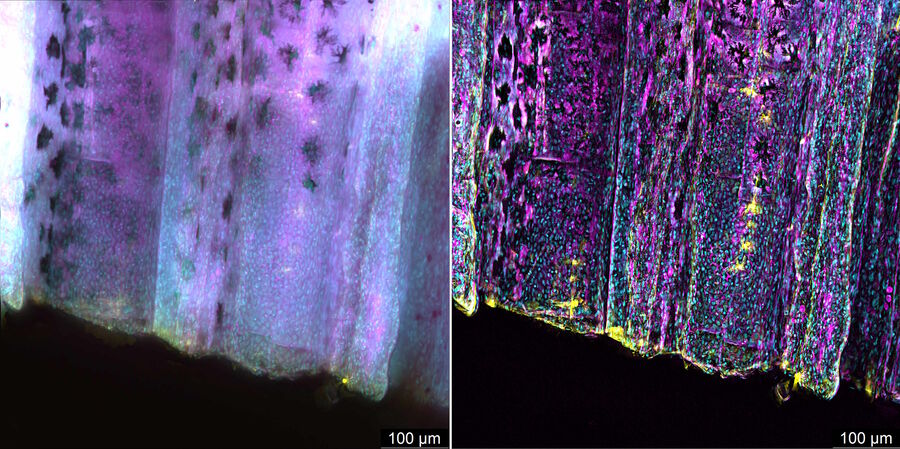
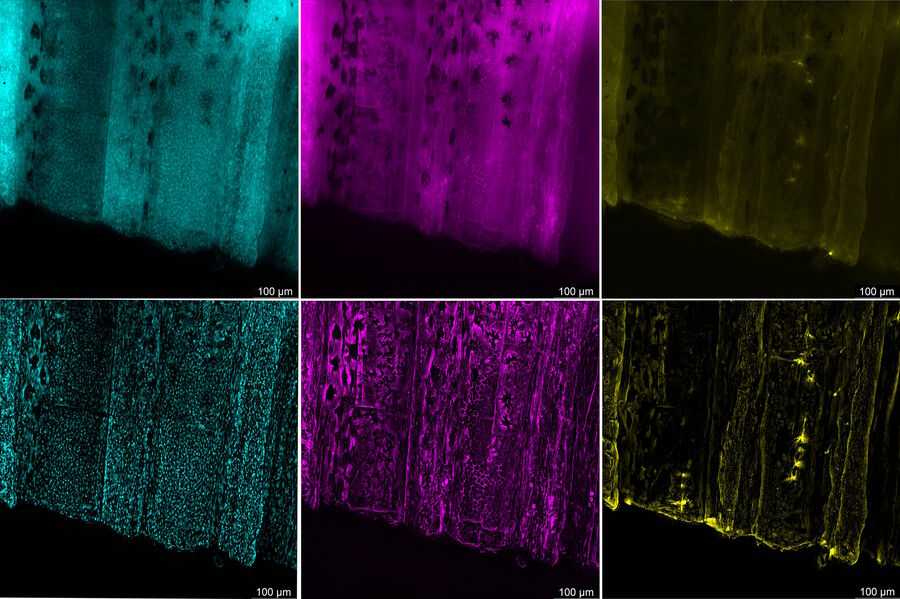
Zebrafish fin tissue immunostained for GFP (yellow), actin (magenta), and nuclei (DAPI, cyan) was used for this study. Specimens were imaged with a THUNDER Imager Live Cell having a 20x, 0.8 NA (numerical aperture) objective. A z-stack of 162 slices every 0.5 µm (total of 80.25 µm) was obtained and processed using large volume (LVCC) or instant computational clearing (ICC) [3,4]. Images were acquired with speeds typical of widefield microscopy. Extended depth of field (EDoF) projections are shown.
Results
With the Thunder Imager Live Cell, the time to acquire a z-stack of 3 channels and 162 slices was just over 1 minute and 10 seconds, allowing a large increase in output compared to confocal microscopy. Additionally, by combining ICC and deconvolution, the background was greatly reduced and fine details more clearly revealed. THUNDER images with enhanced contrast and resolution are seen in comparison to raw widefield ones in figure 1 and 2.
Conclusions
THUNDER images of zebrafish fin, where large volume (LVCC) or instant computational clearing (ICC) were applied, revealed better fluorescence signals of tissue immunostained for GFP, actin, and nuclei compared to conventional widefield microscope images.
References
- Ear, J., Abd El-Hafeez, A.A., Roy, S., Ngo, T., Rajapakse, N., Choi, J., Khandelwal, S., Ghassemian, M., McCaffrey, L., Kufareva, I., Sahoo, D., Ghosh, P. A long isoform of GIV/Girdin contains a PDZ-binding module that regulates localization and G-protein binding, J. Biol. Chem. (2021) vol. 296, 100493, DOI: 10.1016/j.jbc.2021.100493.
- Marivin, A., Maziarz, M., Zhao, J., DiGiacomo, V., Olmos Calvo, I., Mann, E.A., Ear, J., Blanco-Canosa, J.B., Ross, E.M., Ghosh, P., Garcia-Marcos, M. DAPLE protein inhibits nucleotide exchange on Gαs and Gαq via the same motif that activates Gαi, J. Biol. Chem. (2020) vol. 295, iss. 8, pp. 2270-2284, DOI: 10.1074/jbc.RA119.011648.
- J. Schumacher, L. Bertrand, Real Time Images of 3D Specimens with Sharp Contrast Free of Haze: Technology Note THUNDER Imagers: How do they really work? Science Lab (2019) Leica Microsystems.
- L. Felts, V. Kohli, J.M. Marr, J. Schumacher, O. Schlicker, An Introduction to Computational Clearing: A New Method to Remove Out-of-Focus Blur, Science Lab (2020) Leica Microsystems.
Related Articles
-
What are the Challenges in Neuroscience Microscopy?
eBook outlining the visualization of the nervous system using different types of microscopy…
Jun 14, 2023Read article -
Central Nervous System (CNS) Development and Activity in Organisms
This article shows how studying central nervous system (CNS) development in Drosophila-melanogaster…
May 12, 2023Read article -
Going Beyond Deconvolution
Widefield fluorescence microscopy is often used to visualize structures in life science specimens…
Mar 22, 2023Read article