Introduction
The fission yeast Schizosaccharomyces pombe is an ascomycete unicellular fungus that has served as an important model system for studying diverse biological processes that support life, such as the cell cycle, cell morphogenesis, apoptosis, and aging [1]. Its small cell size, however, imposes a significant challenge for live acquisition studies using conventional microscopy where small structures appear blurred by the out-of-focus light. For this reason, traditionally these studies have been carried out using spinning disk microscopy.
Methods
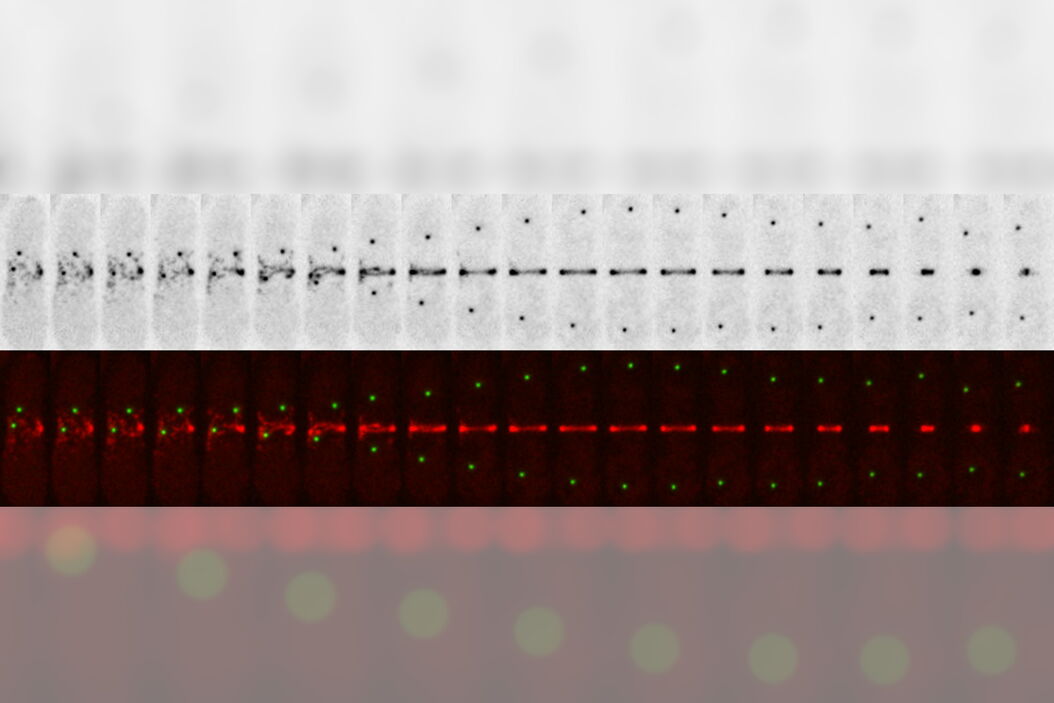
A THUNDER Imager 3D Live Cell has been used with instant computational clearing (ICC) [2,3] to obtain sharp 4D images of live fission yeast cells expressing the ring marker Rlc1-mCherry (myosin II regulatory light chain) and spindle pole body marker Pcp1-GFP (pericentrin/kendrin). To overcome the high autofluorescence levels typical of media commonly used for growing yeast, such as YES, the cells were mounted on top of an agarose coating pad, so as to minimize the amount of medium needed.
Results
The images below demonstrate that the THUNDER Imager 3D Live Cell, together with ICC, is able to provide clear and high-contrast images of fluorescently labeled fission yeast cells.
Cell mounting protocol (by Javier Encinar del Dedo):
- 0.2 g of agarose are dissolved in 10 ml of medium and kept at 65 °C to avoid solidification.
- 15 µl of coating solution are pipetted on top of slides pre-heated at 42 °C, and a second slide is placed on top and kept for 15 minutes.
- After removal of the second slide, the agarose pad is ready to pipette 1.5 µl of cell in early log-phase.
- A coverslip is finally placed on top of the cells before going to the microscope.
- A water immersion lens is recommended to help avoid optical aberrations (63x 1.2 NA) in order to improve the computational clearing.
This short protocol enables fission yeast divisions to be followed for about 1 hour when imaging with a THUNDER Imager 3D Live Cell. Around 16 planes and a minute scale interval were used to achieve low photobleaching and sharp imaging at the subcellular level.
References
- Hayles J, Nurse P. Cold Spring Harb Protoc. 2018 May 1;2018(5). doi: 10.1101/pdb.top079749.
- J. Schumacher, L. Bertrand, THUNDER Technology Note: THUNDER Imagers: How Do They Really Work? Science Lab (2019) Leica Microsystems.
- L. Felts, V. Kohli, J.M. Marr, J. Schumacher, O. Schlicker, An Introduction to Computational Clearing: A New Method to Remove Out-of-Focus Blur, Science Lab (2020) Leica Microsystems.
Related Articles
-
What are the Challenges in Neuroscience Microscopy?
eBook outlining the visualization of the nervous system using different types of microscopy…
Jun 14, 2023Read article -
Central Nervous System (CNS) Development and Activity in Organisms
This article shows how studying central nervous system (CNS) development in Drosophila-melanogaster…
May 12, 2023Read article -
Going Beyond Deconvolution
Widefield fluorescence microscopy is often used to visualize structures in life science specimens…
Mar 22, 2023Read article