Introduction
Viral entry into a host cell requires coordination of both viral and cellular proteins in a precise order to trigger membrane fusion. Here we present two examples from the field of HIV research to demonstrate how advanced confocal microscopy techniques enable time-resolved stoichiometric analysis of HIV fusion events in live cells [1] and how fluorescence lifetime imaging (FLIM) facilitates the understanding of the infection changes to the host. This result is achieved with molecular reporters that give a fluorescent readout of physiological changes within the cell. Today’s new generation of confocal microscopes—with the ability to perform lifetime-based imaging—has facilitated use of multiple fluorescent biosensors in the same experiment. Application of FLIM to Förster Resonance Energy Transfer (FRET) determination also allowed the collection of more quantitative data. This approach has been highly informative in elucidating the role of cellular metabolism in viral susceptibility, as well as the subsequent post-infection effects on cellular metabolism [2].
Time-resolved stoichiometry of the HIV fusion reaction in live cells
Getting a handle on viral entry mechanisms
HIV entry into immune cells, such as macrophages and T cells, requires a complex and coordinated cellular response that is initiated by interaction of the viral particle with cell surface receptors. Immature HIV particles are not able to enter a host cell because of the rigidity of their glycoprotein spikes [3]. Before becoming infective, viral particles require Gag proteins to be cleaved. This step enables a structural reorganization that results in fluidity of the viral receptor binding protein Env at the viral membrane. Once the Env proteins can move freely, they are able to form Env trimers in the membrane, thus providing the functional membrane cluster needed for host membrane fusion (for details see the review Jakobsdottir et al. 2017) [4]. This interplay between the viral glycoprotein and the host receptors is the key initiation step that determines an effective infection event. A more complete understanding of the molecular mechanics of this infection process is thus highly desirable, as it could be exploited for the development of novel intervention strategies to prevent viral infections.
Gaining insight with quantitative photon counting and fluorescence lifetime information
Fluorescence Lifetime Imaging Microscopy (FLIM/FALCON [5]) and photon counting-based advance analysis are all powerful techniques for measuring molecular dynamics and mechanisms. Recent advances in photon counting introduced with the STELLARIS platform increase the ability for further discrimination of photon events and improve the fidelity of the measured fluorescence [6]. Taking advantage of fluorescence-lifetime based information to distinguish multiple biosensors in the same experiment is an effective means of elucidating the complex, multi-stage, mechanisms involved in viral entry. Recent advances in confocal microscope technology present in the STELLARIS platform allow fluorescence lifetime information to be captured on a confocal microscope [5],[7]. FLIM can also be used to measure FRET which is an important technique for understanding dynamic spatial relationships between molecules. FLIM-FRET is a gold standard technique, because it provides more robust and quantitative data than other FRET methods, such as sensitized emission or photobleaching.
Tracking single particles and molecules in real time
When studying viral entry, a critical goal is to understand how the virus particle physically interacts with the host cell. Microscopy techniques have now advanced to the point where it is possible to analyze these interactions at the molecular level. Within any given illumination volume in a fluorescence microscopy experiment, the maximum fluorescence intensity of a given molecule is directly proportional to the total number of that molecule [8]. In addition, the corresponding amplitude of the intensity fluctuations will vary depending on whether the molecules are in a monomeric or a dimeric state [9]. These fluctuations require time-resolved microscopy techniques in order to accurately quantify them. Using this information, it is possible to track single viral particles as well as single molecules on the cell surface to get an accurate picture of the stoichiometry of virus receptor and co-receptor binding.
There are three microscopy methods in particular that have been key to the study of viruses in real time:
- Single viral particle tracking (SVPT) – individual viral particles are tracked and their spatiotemporal interactions with host cells receptors are monitored. Viral particle tracking is used as the anchoring point for the cellular analysis. It also allows for the cellular signal, which does not correlate with particle tracking, to be used as an internal control measure.
- Number & Brightness (N&B) [6]-[8] – time-dependent fluctuation of fluorescent signals can be monitored to determine the oligomeric state of a given protein in real time using fluorescence correlation spectroscopy ( FCS ). This information is used to assess the number of each receptor involved in the virus-host interaction.
- Cross-correlation N&B (ccN&B) [10],[12] – positive correlation of signals from two fluorescently tagged proteins is a hallmark of protein-protein interaction or protein complex formation. Cross-correlation of N&B readouts enables tracking of whether host receptors interact with each other during the infection, also giving the stoichiometry. Such interactions are the initiation points of virus-induced signaling cascades.
Building a detailed picture of HIV entry using quantitative fluorescence intensity-based signals
A study by Iliopolou et al. published in Nature Structural and Molecular Biology used these methods to reveal the dynamic process by which HIV fuses to the host cell membrane [1]. The HIV-1 Env protein binds primarily to a single CD4 molecule and a CXCR4 molecule, the receptor and co-receptor interacting with each other and the HIV Env trimer throughout the fusion process. Shortly after this initial complex formation, a CXCR4 dimerization is observed. It is worth highlighting that the HIV co-receptors are GPCRs, and such a dimerization step could serve as the trigger point for cellular signals needed to facilitate membrane reorganization and fusion from the host side. The CXCR4 dimerization” acts as the anchor for the HIV particle, as more CD4 molecules are recruited to form a large complex on the cell surface. Additional rearrangements of this protein complex then favor stabilization of the reaction and displacement of the co-receptors to allow for the six-helix-bundle formation needed for fusion of the viral particle and entry into the host cell [1]. The work by Iliopolou et al. showed the specific spatiotemporal mechanisms for both X4 and R5 tropic viruses. Their data suggests a 3-step mechanism for the formation of the prefusion complex. This time-resolved view, together with different HIV glycoprotein crystal structures published to date, has enabled the group to build a detailed picture of the mechanics at play during HIV entry [1].
Role of cellular metabolism in HIV infection
Another important aspect of viral entry is its heterogeneous nature. Even when studying cultures of genetically identical cells, it is often observed that not all cells in the population are susceptible to a given infectious pathogen at the same time. This phenomenon has been observed with many different viral pathogens, including HIV-1. Together with studies that show a clear link between metabolic pathways and important T-cell functions [13],[14], these observations led to the hypothesis that a cell’s metabolic state has an effect on viral entry [2]. When 2‑Deoxyglucose was used to disrupt cellular metabolism, the percentage of virally infected cells also decreased.
Optical biosensors and lifetime imaging enable deeper exploration of molecular mechanisms in viral entry
Discovering this correlation between cellular metabolism and viral susceptibility was only the tip of the iceberg. It was not until the availability of advanced imaging techniques that it became possible to decipher the underlying mechanisms. One central question remained unresolved: does the virus alter cell metabolism, or does heterogeneity in cell metabolism drive infection hotspots [2]. To address this question, researchers have made use of a variety of fluorescent probes and optical biosensors to detect and correlate different parameters important for viral entry, such as metabolic activity, viral fusion events, and localized membrane tension. All these biosensors monitored using time resolved techniques which is essential for gaining insight into dynamic biological processes, such as viral entry. Lifetime imaging allows more than one biosensor to be used in the same experiment as well as generating more robust and quantifiable data in the case of FLIM-FRET [8].
Metabolic activity sensors
General metabolic biosensors such as Perceval using GFP [15] can give optical readouts of the cellular energy status by measuring the ATP/ADP ratio. The amount of ATP present reflects the energy stored in a cell, whereas ADP levels rise as energy is consumed during various metabolic reactions. The ratio of the two, therefore, gives a good indication of metabolic activity in the cell. Using these assays in combination with a fluorescently labeled virus particle tracking probe, it is easy to see the correlation between viral particles and metabolically active cells. This method is relatively broad and cannot provide detailed information about how viral entry and metabolism are interlinked.
Viral fusion biosensor (iBLAM)
More details about the interactions of viral particles and the host cells can be uncovered by using reporters that give an optical readout when fusion takes place. A good example is the iBlaM reporter where a beta-lactamase-vpr chimeric protein is encapsulated in the HIV capsid. If a fusion event occurs, the beta-lactamase-vpr enters the cell and cleaves the dye CCF2 causing it to emit a different wavelength of light. This readout can also benefit from fluorescence lifetime measurements [16].
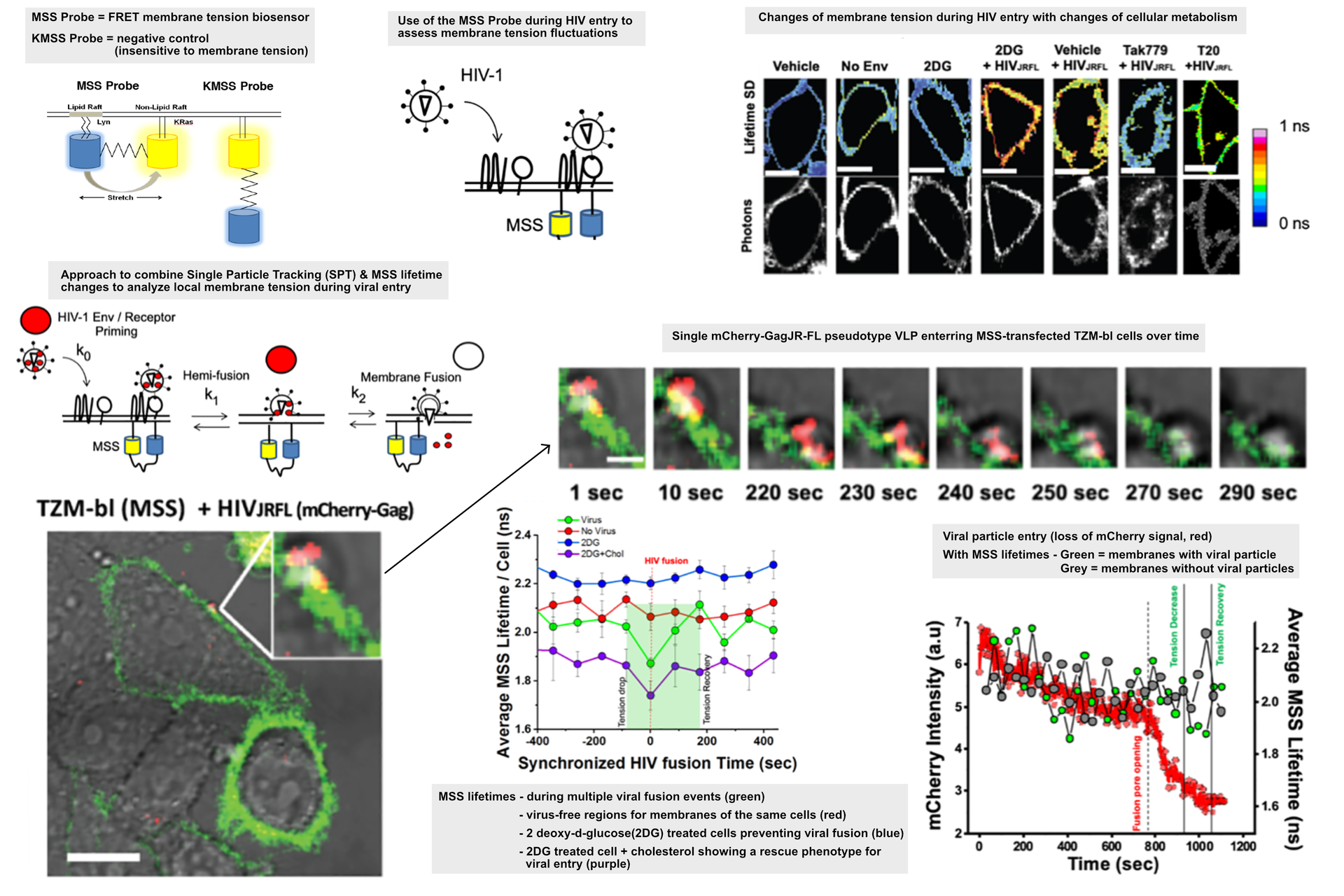
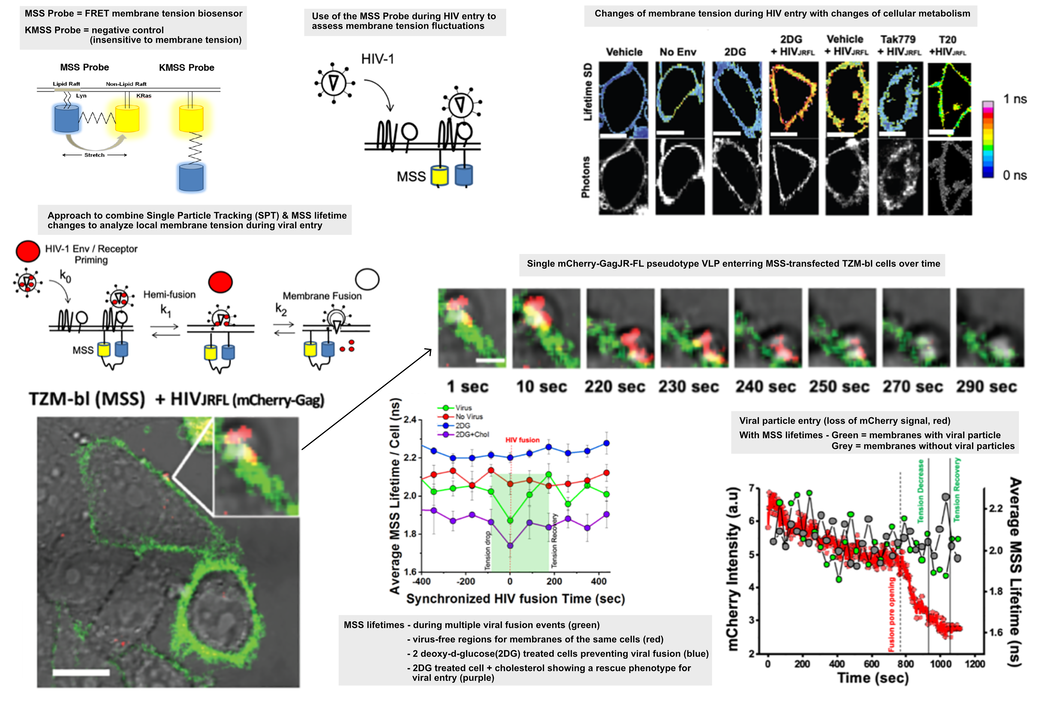
Membrane tension biosensors
There are many innovative biosensor systems that can give time resolved information about the state of the host cell. When these are introduced into an infection model, they can quickly generate novel insights into the infection process. A FRET based biosensor was used to give an optical, quantitative readout for the localized tension in the cell membrane [17] where the HIV particles were binding. The biosensor is based on a eCFP-Ypet pair linked to a Lyn and KRas sequence, respectively [17]. A change in the fluorescence lifetime signal gives an indication of how much the fluorescent protein could be stretched between ordered and disordered membrane motifs which is in turn correlated to membrane tension. The data showed that as HIV particles were binding, there was a localized decrease in the membrane tension. This drop could be prevented by treating the cells with 2 deoxy-glucose which also prevented viral entry by inhibiting glycolysis and reducing the amount of available cholesterol at the membrane. In this case, there is a decrease in membrane order and increased tension which ultimately prevents viral entry [2].
Conclusion
The studies cited here highlight some of the major inroads that have been made in HIV research and the same techniques can be applied to the study of dynamic processes at different stages of the HIV life cycle or even other viruses. Discussed here were fluorescent probes and biosensors used to study metabolism and membrane tension which are both relevant for viral entry studies. However, there are many more biosensors available for a plethora of cellular processes. A recent database has been published that can provide a great starting point for experimental design [18]. Finding the right one will depend on the hypothesis to be tested with the experiment. It is also important to consider how well the biosensors fit with your confocal platform. The ability to combine multiple biosensors in the same experiment, as well as the ability to use lifetime-based information from FRET and other fluorescence-based systems, allowed for new insights to be gained from complex, multi-step host-virus interactions.
Modern confocal platforms with their sensitive photon counting detectors enable the visualization and quantification of single molecular events and the study of rapid changes at the molecular level in real time [1],[6],[10]. Advances in fluorescence lifetime imaging techniques have allowed researchers to benefit even more from optical biosensors and gain a much deeper understanding of the molecular processes driving HIV cell entry and infection [2],[5]. Ultimately, this technique will help researchers develop new intervention strategies that target the fundamental aspects of HIV infection that are necessary for vaccines. In addition, such approaches can be generalized for the study of other known and emerging viral pathogens.