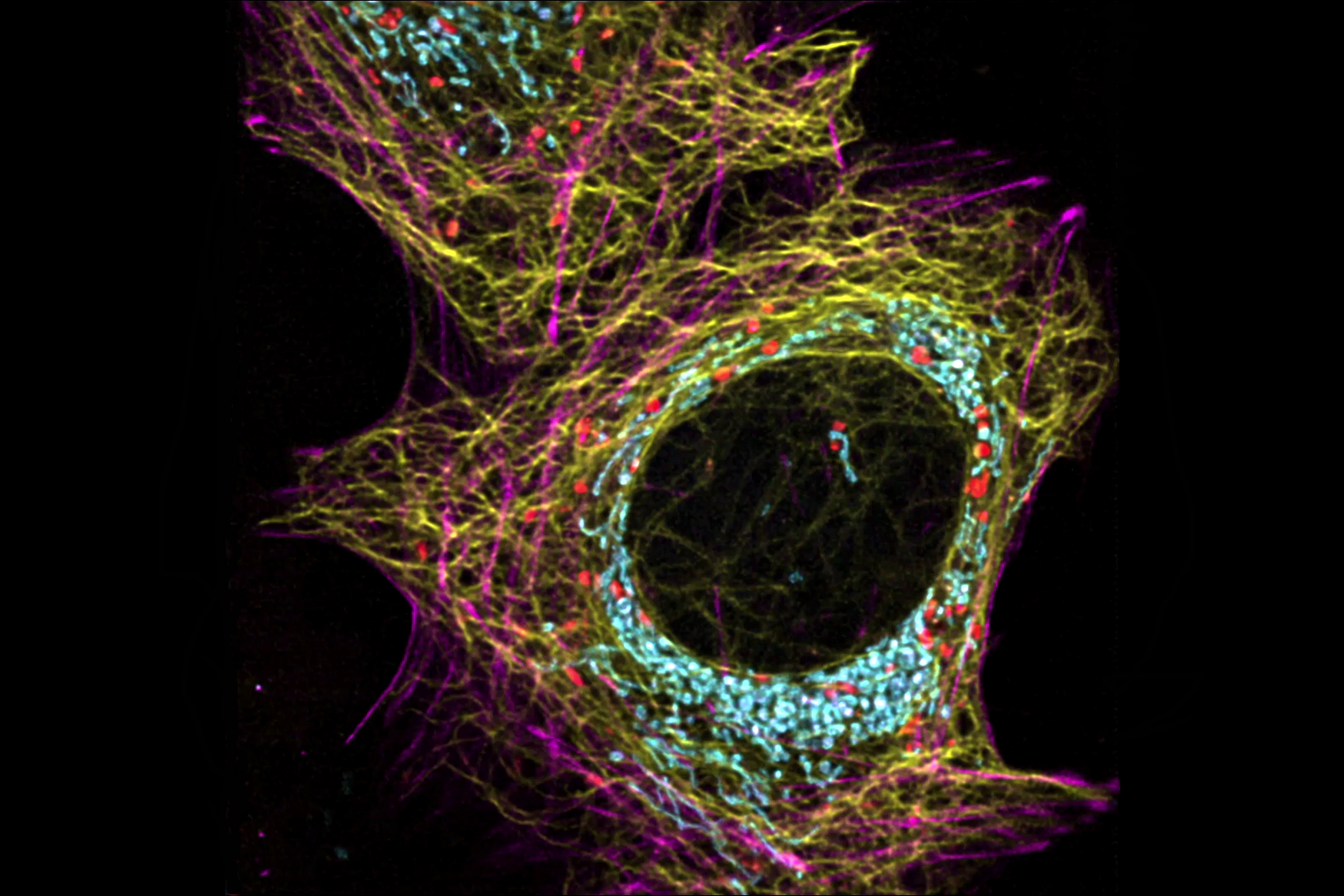
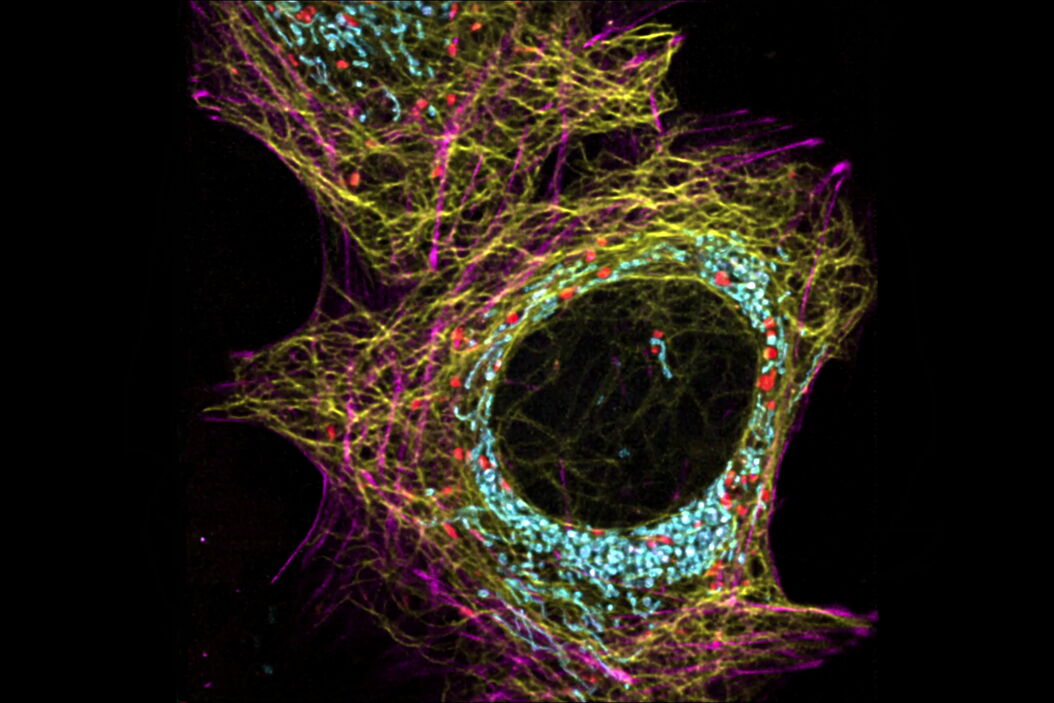
Introduction: An excellent standard for live-cell 3D imaging
Life is complex, dynamic and happens in 3D. The STELLARIS confocal platform is a great choice for capturing these hallmarks of life. Live-cell imaging, with STELLARIS enables you to capture the complex dynamics of the subcellular machinery and its microenvironment over time and with high 3D resolution.
STELLARIS hardware for gentle and fast live-cell imaging
Confocal microscopy is generally considered a gentle, non-invasive method and well suited for live-cell studies. This fact is particularly true for STELLARIS. On the one hand, STELLARIS comes with a white light laser (WLL) that allows the matching of fluorophore excitation spectra to minimize adverse effects, such as phototoxicity. On the other hand, the sensitive Power HyD detector family enables you to faithfully detect even the faintest signals over a wide spectral range. Finally, employing the resonant scanner, STELLARIS is not only gentle to the sample, but also allows for capturing dynamic changes in live specimens with high temporal resolution.
Challenges of live-cell imaging
As researcher, you set the goal to discover novel insights and, thus, often push the limits of your imaging experiments. Accordingly, you may confront these challenges:
- Can I get better temporal resolution to record neural activity even more accurately?
- Can I visualize additional markers to get even more context from the immunological synapse?
- Can I prolong my experiment to follow the behavior of cancer stem cells even longer?
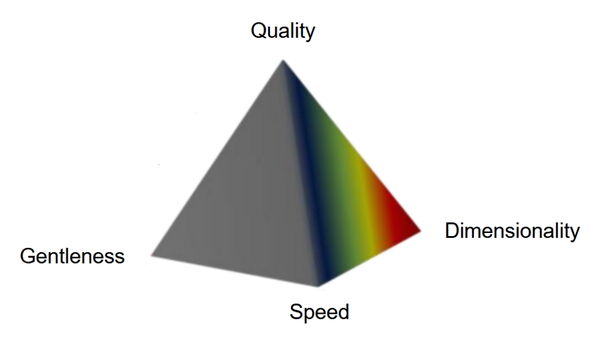
These demands will inevitably require some trade-offs. The seemingly mutually exclusive limitations can be visualized with the imaging pyramid: gentleness, speed, image quality, and dimensionality. Consequently, tools which mitigate these trade-offs are highly sought after.
Dynamic Signal Enhancement (DSE) for increasing the signal-to-noise ratio (SNR)
In practice, the crucial trade-off for live-cell imaging is frequently between acquisition speed and image quality, because experimental gentleness and dimensionality often have certain requirements. Accordingly, since capturing the dynamics of living samples requires higher frame rates, live-cell imaging experiments commonly have a lower image quality than fixed samples due to less time for the detectors to capture photons from the specimen.
With STELLARIS, we introduce Dynamic Signal Enhancement (DSE) for real-time denoising of live-cell imaging series. DSE uses a rolling average calculation as its core algorithm. The basic idea of a rolling window is that noise between adjacent imaging frames is random while the biological signal is conserved. Accordingly, by averaging over several adjacent raw imaging frames with a rolling window, the relevant biological signal is accentuated while the unwanted noise is “averaged out”. In more expert terms, the DSE increases the signal-to-noise ratio (SNR) and ultimately leads to better image quality.
Other “simple averaging” methods, i.e., averaging several raw images to output a single denoised image, reduce the temporal resolution by the averaging factor. For example, if you were averaging five raw image frames, the temporal resolution of the output image would be decreased by a factor of five. In contrast, DSE keeps the raw image frames and slides a fixed size averaging window over the entire raw image series. As result, the number of image frames and temporal resolution are identical between raw and DSE-processed imaging data.
Aivia powers the Dynamic Signal Enhancement: Better temporal dynamics and better signal-to-noise ratio at the same time
However, since we integrate over several raw data frames with the rolling average, the temporal dynamics could be reduced when applying DSE. For example, when using an unfavorable high number of frames for the rolling window, dynamic structures can begin to smear and show signs of doubled structures (“ghosting”). Accordingly, using a rolling window for denoising has its own trade-off: When averaging over more frames, the SNR gets better, but the temporal dynamics of fast processes are reduced. When averaging over less frames, the SNR is worse, but the temporal dynamics of fast processes are better preserved.
Here, the new DSE option Dynamic Signal Enhancement powered by Aivia helps to boost your live-cell imaging results. With a single click, Dynamic Signal Enhancement powered by Aivia uses AI to interpolate frames. Using this additional information for rolling window calculation, DSE can now reach a higher SNR and better temporal dynamics at the same time.
In the animation from above, we have virtually doubled the temporal dynamics of the sample by only using three acquired raw images for the rolling window. At the same time, we still average over a total of five frames by using two additional interpolated frames for the rolling window calculation. While it may seem intuitive that this should lead to the same SNR as for DSE with five raw frames and no Dynamic Signal Enhancement powered by Aivia, in practice, we saw a further improvement of the SNR. This observation can be explained by how the Dynamic Signal Enhancement powered by Aivia deep learning network works: Dynamic Signal Enhancement powered by Aivia also acts as a filter by interpolating pixels, which are consistent over frames (i.e., biological signal), rather than noise.
It is important to note that the number of frames you get as a result of Dynamic Signal Enhancement powered by Aivia does not change. This number is identical to the number of raw data frames used as your input. So even though Dynamic Signal Enhancement powered by Aivia may have “doubled” the number of frames used as your input for DSE, these frames are only transient, resulting in a higher SNR and better temporal dynamics for your live-cell experiment when using STELLARIS.
Combine with LIGHTNING
The STELLARIS software offers even more tools to improve your live-cell imaging experiment. The LIGHTNING detection concept is based on a fully automated, adaptive deconvolution method which you can easily combine with Dynamic Signal Enhancement powered by Aivia in a single processing step. In this way, your live-cell imaging series gets improved in multiple ways – not only the noise gets removed but also the contrast is enhanced, and the spatial resolution further refined. Thus, by combining DSE and LIGHTNING, we can mitigate the restrictions of the imaging pyramid even more.
Conclusion
If you’re a scientist performing live-cell 3D imaging, now you can benefit from more advanced data acquisition during your experiments and collect more accurate and reliable results for publication. You just need to take advantage of the power of AI with Leica Microsystems’ brand new Dynamic Signal Enhancement powered by Aivia software package for the STELLARIS confocal platform.