Abstract
Some of the most prevalent neurodegenerative diseases, including Alzheimer’s Disease (AD) and Parkinson’s Disease, are characterized by the appearance of localized pathological structures in affected brain tissues and a corresponding progression of increasingly severe neurological symptoms. A common feauture of these tissue pathologies is that they typically involve complex aggregates of aberrantly folded proteins and associated lipid-rich structures [1–4]. Despite decades of intense research, however, the molecular mechanisms of the respective diseases remain incompletely understood and it remains unclear whether the observed tissue pathologies are a cause or consequence of the disease.
Therefore, methods that can provide increasingly detailed biochemical and biophysical characterizations of pathological structures in brain tissues are in high demand. In a recent preprint [4], we demonstrated the potential of Stimulated Raman Scattering (SRS) microscopy to provide unique insights into the interplay between pathological Amyloid-β (Aβ) aggregates and relevant lipid species in affected brain tissues from a mouse model of AD. In particular, we demonstrated a two-color SRS procedure that enables a clean, label-free visualization of pathological Aβ aggregates amongst the vast healthy protein content of the brain. Furthermore, SRS spectroscopic imaging allowed a detailed spatially-resolved profiling of brain lipids and demonstrated a clear differential localization of several relevant classes of lipids in healthy and diseased brain structures.
In the following, we provide a concise summary of our results. For further details, feel free to refer to the complete manuscript [4] which is publicly available.
Results
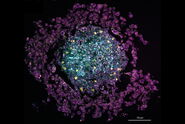
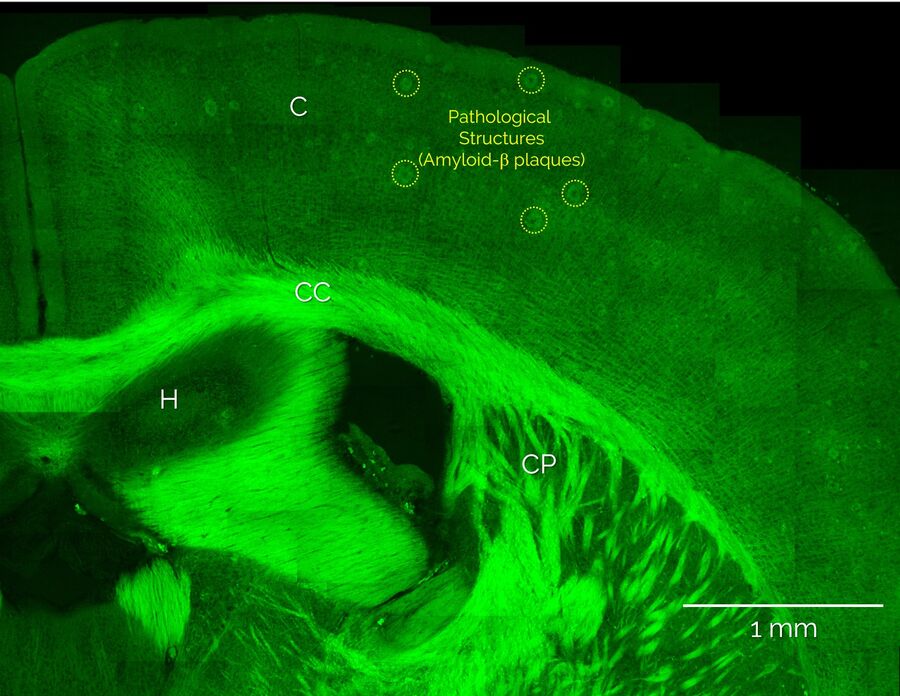
Unlabeled brain slices from mice carrying an amyloid precursor protein and a presenilin 1 mutation that exhibit AD-like pathology [5,6], as well as from healthy control mice, were provided by our collaborators at the German Center for Neurodegenerative Diseases, DZNE (Bonn Germany). Coherent Anti-Stokes Raman scattering (CARS) microscopy and stimulated Raman scattering (SRS) was performed using the SP8 CARS microscope from Leica Microsystems with SRS add-on. Additional nonlinear optical signals (2-photon autofluorescence, second-harmonic generation) were acquired in parallel. Figure 1 shows a large-area overview SRS image of the cortex and hippocampus areas and intertwining white matter regions. Here, the distribution of lipids is visualized by SRS signals from the CH2 stretch vibrations at 2850 cm-1. Hence, lipid-rich structures appear bright, such as the white matter bundles of the corpus callosum (CC) and the individual myelinated neurons that are found throughout the cortex (C), whereas structures devoid of lipids, such as neuronal cell nuclei, appear dark. In addition, Figure 1 also shows large numbers of localized lipid-rich structures in the cortex that are only seen in diseased brain slices. In the following, we show that these structures correspond to pathological lipid deposits associated with Amyloid-b plaques.
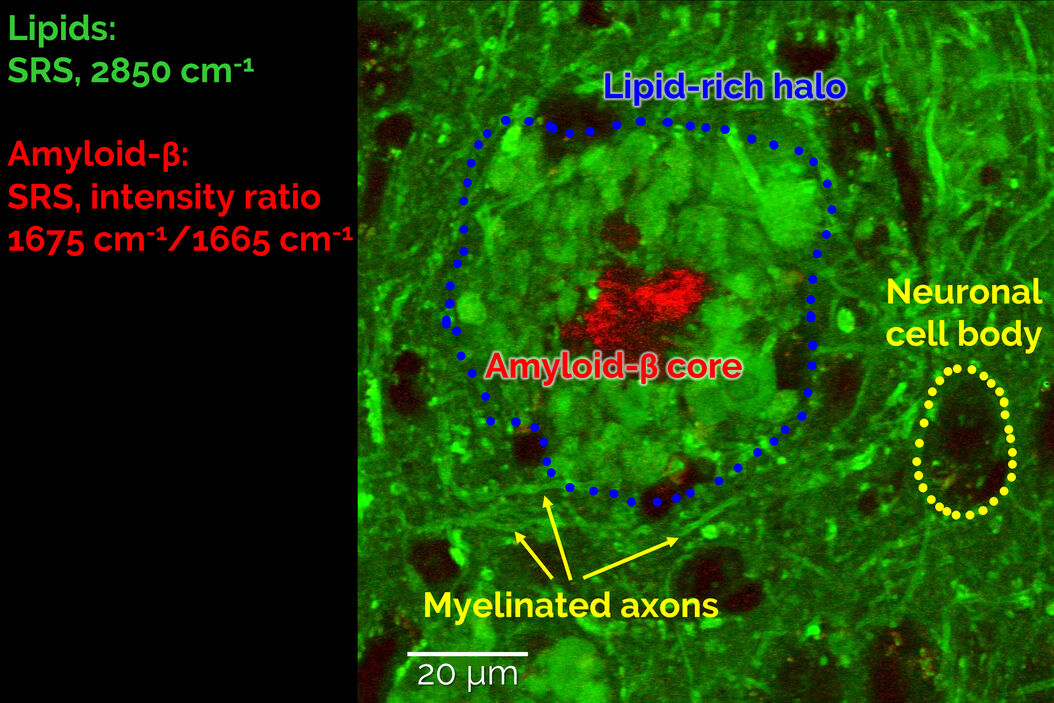
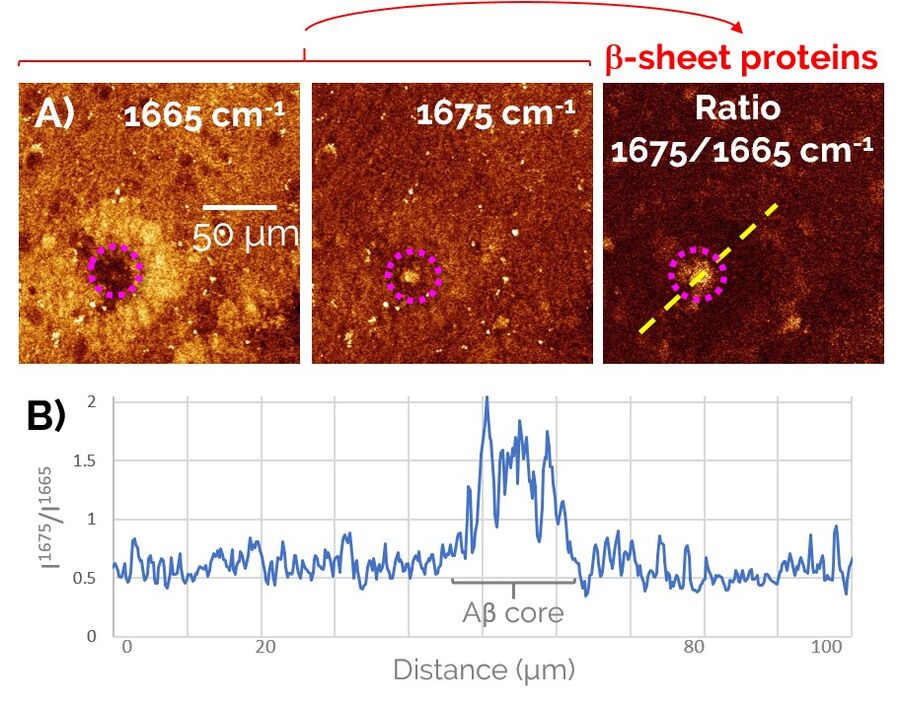
In order to study Alzheimer’s pathology without a need for fluorescent labeling, first we devised a two-color SRS prcedure to specifically visualize pathological Aβ aggregates amongst the healthy protein and lipid content of the brain (Figure 2). To do so, we exploited a frequency-shift of the Amide I mode that arises from the misfolding of peptides into a β-sheet configuration, as is the case for the aggregated forms of Aβ peptides. By taking the ratio of the SRS contrast at 1675 cm-1 (peak of the “misfolded” Amide I mode) and 1665 cm-1 (“regular” Amide I mode), we were able to obtain a clean signature of the Aβ aggregates in brain tissues (right panel of Figure 2).
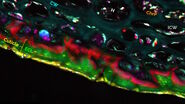
A combination of this ratiometric Amyloid-β signature with the known contrast from lipids at 2850cm-1 allowed us to study the co-localization behaviour of lipids with pathological Amyloid-β aggregates. As shown in Figure 3, we found that the cores of the Aβ aggregates were consistently surrounded by prominent halo-like arrangements of lipid-rich deposits. Healthy brain slices neither showed Aβ aggregates nor substantial localized lipid deposits throughout the cortex.
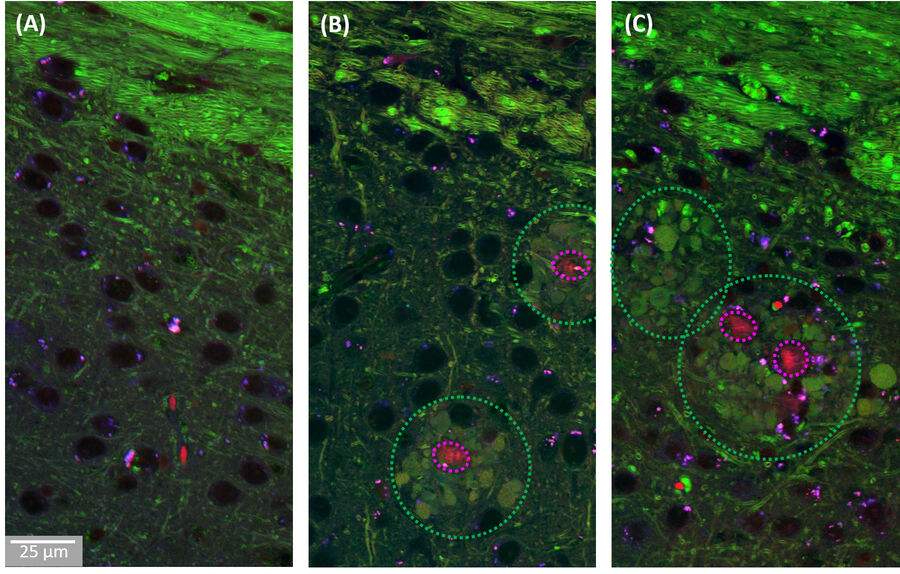
The observed lipid deposits are morphologically complex, consisting of multiple distinct features ranging in size from a few to ten micrometers. Some of these structures show cell-like appearances, with convex shapes and a round central core containing lower lipid concentrations, resembling a nucleus. Interestingly, earlier studies using fluorescence microscopy have shown that microglia/macrophages can be recruited to Aβ plaques.
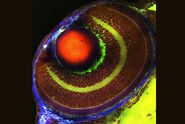
The observed association of lipid structures with Aβ aggregates is particularly intriguing, as recent hypotheses suggest that certain classes of lipids may play a role in the progression of AD by affecting the transport of neurotoxic Aβ species within the brain. Therefore, we decided to study the biochemical composition of these lipid deposits in more detail using SRS spectroscopic imaging, also known as hyperspectral SRS imaging. Representative SRS images and spectra are shown in Figure 4.
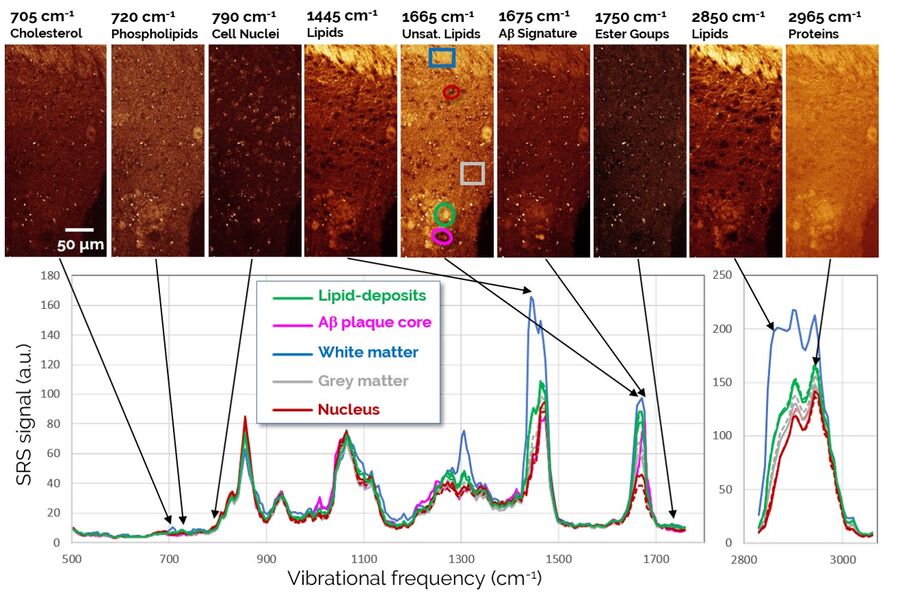
The SRS spectra of pathological lipid deposits are shown in green in Figure 4. For comparison, spectra of lipid-rich healthy white matter, protein-rich gray matter, and neuronal nuclei are shown as well. In particular, the fingerprint region (1800-500 cm-1) provides rich spectral content enabling a detailed biochemical characterization of the tissues. Both white matter and pathological lipid structures show high concentrations of unsaturated lipids (1665 cm-1), but very little signs of lipid oxidation (signals from ester groups at 1750 cm-1 are largely absent). Further signatures in the 900-1400 cm-1 range would allow an even more detailed fingerprinting of the lipids, however, this analysis is complicated by the background from protein signals that is also present in this range. Surprisingly, the apparently weak signals in the region around 700 cm-1 again contain significant information on several highly relevant lipid species. This region is examined in detail in Figure 5.
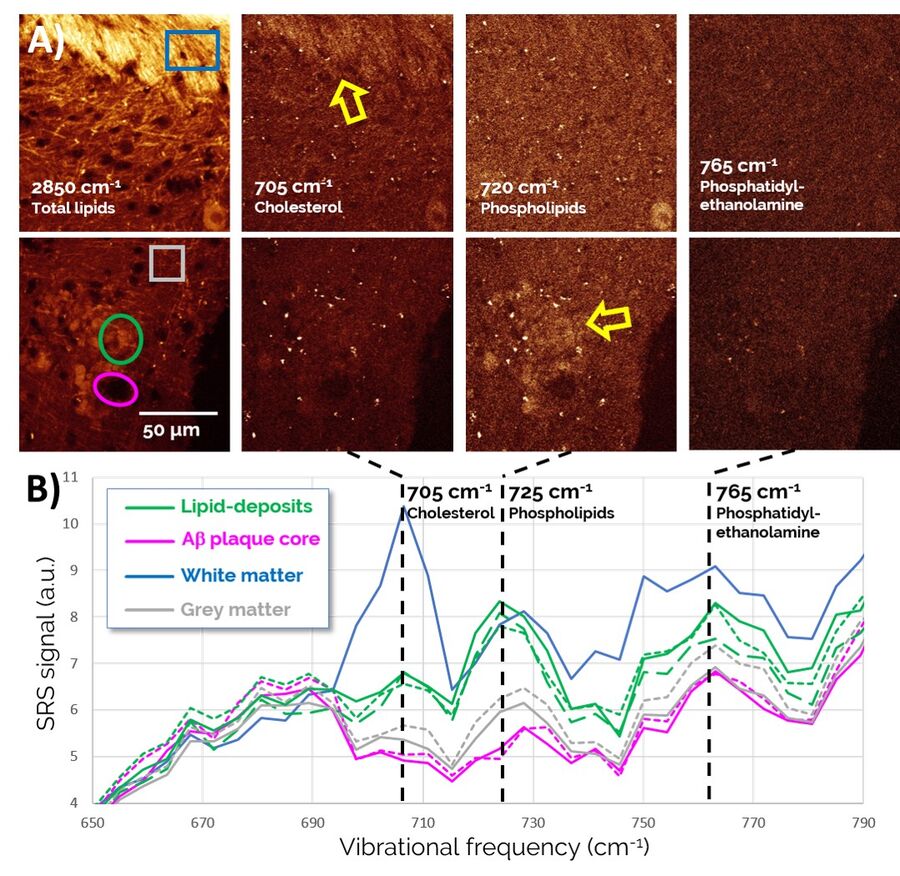
Zoomed-in spectra of the 650-790 cm-1 region, depicted in Figure 5, reveal a number of lipid-specific resonances: The sterol ring breathing mode, found at 705 cm-1, reveals that Cholesterol is present in high concentrations in healthy white matter (top of the images) as expected, but does not show much specific localization to Aβ plaque-associated lipid deposits (bottom of images). By contrast, membrane phospholipids, such as Phosphatidylcholine and Sphingomyelin (visualized through a vibration of the choline group at 725 cm-1), are found highly enriched in plaque-associated lipid deposits. Phosphatidylethanolamine (765 cm-1), another prominent lipid in the brain, is found more diffusely throughout the entire tissue region and does not appear to play an obvious role in AD pathology.
Discussion
Interactions of lipids with pathological Aβ structures are under active investigation for their relevance to AD disease progression. The reason for this activity is because lipids are thought to have the capacity to extract small Aβ oligomers from larger aggregates in a detergent-like fashion, thereby promoting neurotoxicity which is thought to arise exclusively from small Aβ oligomers. Furthermore, an aberrant metabolism or transport of several major lipid classes, including cholesterol, is known to be associated with AD.
Our spectroscopic analysis of lipid deposits in the vicinity of Aβ-plaques reveals a clear differential localization of important lipid species in these pathological structures: Plaque-associated deposits are found to be rich in the membrane lipids phosphatidylcholine and sphingomyelin, but much lower in cholesterol than nearby healthy lipid-rich brain structures. These observations provide important clues as to the source of plaque-associated lipids, suggesting that the membranes of nearby neurites are a more likely source than potential alternative mechanisms associated with an aberrant cholesterol transport. Some of the lipid structures appear to bear a morphological resemblance to microglia/macrophages. Therefore, it will be an interesting future avenue of research to investigate whether the brain’s innate immune system could play a role in recruiting lipid species to sites of neurodegeneration and whether such mechanisms could be exploited to fight the disease.
Taken together, our results highlight the potential of SRS to contribute more broadly to a deeper understanding of neurodegenerative diseases [1–4,7,8]. In the future, we anticipate that the spectroscopic imaging capabilities of SRS will enable similar applications in many areas of the life sciences and medicine. These areas range from fundamental research in cell and tissue biology to preclinical research on disease mechanisms to applications in histopathology and diagnostic imaging as well as the identification of novel biomarkers for a wide range of diseases.
Further Reading
- Kiskis, J. et al. Plaque-associated lipids in Alzheimer’s diseased brain tissue visualized by nonlinear microscopy. Sci. Rep. 5, 1–9 (2015).
- Shahmoradian, S. H. et al. Lewy pathology in Parkinson’s disease consists of crowded organelles and lipid membranes. Nat. Neurosci. 22, 1099–1109 (2019).
- Ji, M. et al. Label-free imaging of amyloid plaques in Alzheimer’s disease with stimulated Raman scattering microscopy. Sci. Adv. 4, eaat7715 (2018).
- Schweikhard, V., Baral, A., Krishnamachari, V., Hay, W. C. & Fuhrmann, M. Label-free characterization of Amyloid-β-plaques and associated lipids in brain tissues using stimulated Raman scattering microscopy. bioRxiv 789248 (2019). doi:10.1101/789248
- Jankowsky, J. L. et al. APP processing and amyloid deposition in mice haplo-insufficient for presenilin 1. Neurobiol. Aging 25, 885–892 (2004).
- Schmid, L. C. et al. Dysfunction of Somatostatin-Positive Interneurons Associated with Memory Deficits in an Alzheimer’s Disease Model. Neuron 92, 114–125 (2016).
- Poon, K. W. C. et al. Lipid biochemical changes detected in normal appearing white matter of chronic multiple sclerosis by spectral coherent Raman imaging. Chem. Sci. 9, 1586–1595 (2018).
- Tian, F. et al. Monitoring peripheral nerve degeneration in ALS by label-free stimulated Raman scattering imaging. Nat. Commun. 7, 13283 (2016).