The principle of GSDIM microscopy
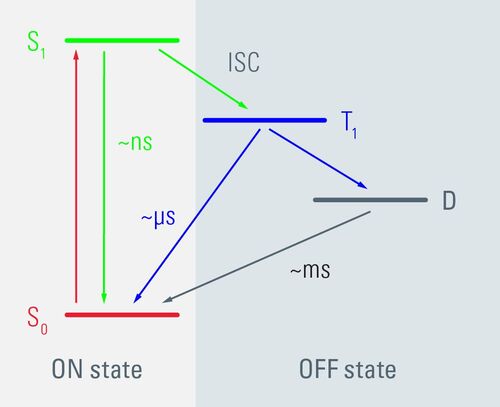
In conventional fluorescence microscopy, delocalized π electrons in a fluorescent dye are transferred from a ground state S0 to an excited state S1. As they oscillate back into the ground state S0, they emit fluorescent light. The GSDIM technique lowers the number of electrons that are involved in this oscillation cycle by switching most of the fluorescent dye to a dark state (Figure 1) (Fölling et al., 2008, Bierwagen et al. 2010, Testa et al., 2010). This reduces the number of excitable fluorophores until single molecules can be detected. Single molecules then return spontaneously from the dark state to an excitable ground state and fluoresce, while others are deactivated again by switching to the dark state. As a result, the fluorophore molecules light up or "blink" in the specimen. The exact position of single fluorophores can be determined by an algorithm. The position information of all the fluorophores is collected in several thousands of separate images, whose coordinates are used to compute a super-resolution GSDIM image. The structures shown in the GSDIM image therefore originate in a similar way to the pointillistic style of painting.
Fig. 1: Schematic representation of the GSDIM method based on a simplified Jablonski diagram. Delocalized π electrons in fluorophores can be, for instance, in the ground state S0, in the excited state S1 (both so-called ON states) or in a triplet or radical dark state (both OFF states). When fluorescent light is emitted, electrons circulate between the ground and the excited state. Unlike these ON states, fluorophores in the OFF state are not able to emit light. These OFF states are usually of long lifetime, but they are difficult to attain, as an inter system crossing is required. By setting the right ambient conditions in the embedding medium and through the clever choice of standard fluorophores for immunofluorescence, it is possible to reversibly switch off fluorophores by exciting them with an extreme light intensity. When enough molecules are in the OFF state, it is possible to detect individual molecules in the sample.
The primary cilium, an unconventional organelle for the polar organization of epithelial cells
Primary cilia are cellular microtubuli-based structures on the apical surface of polarized epithelial cells of the human body. This form of monocilia is found, for example, on tubuli-coated epithelial cells of the kidney, the biliary and the pancreatic duct. Here they play a key role as mechanical sensors – for instance, they determine the composition of fluids or their flow rate (Fliegauf et al., 2007). At the same time, they are involved in maintaining cell polarity, as has been shown with Madin Darby canine kidney (MDCK) cells (Bacallao et al., 1989). Primary cilia are formed during interphase of the cell cycle from a plasma membrane-associated basis about 250 nm wide called the basal body, i.e. the axoneme is formed on the basis of this structure (Singla et al., 2006). The basic framework of the basal body consists of nine microtubule dimers (9+0). Various centrins are located here that are regarded as essential components of the basal body. These proteins belong to the calmodulin family and have an EF hand motif with which they are able to bind Ca2+ (Salisbury, 2007). The conformation change induced by the Ca2+ binding regulates the interaction of centrin with other proteins such as tubulin or galectin-3. There are four different types of centrin in the murine and human organism. Centrin-2 and -3 are expressed ubiquitously and accumulate in the centriolar and pericentriolar areas or in the basal body of the cell. Centrin-1, on the other hand, is only found in germ cells and the retina, while centrin-4 is only found in fully differentiated cells (Salisbury, 2007).
Disorders in ciliogenesis or the function of the cilium are the cause of various diseases such as retina degeneration, polycystic kidney disease, hydrocephalus or BBS (Bardet-Biedl syndrome).
Protein components of the basal body imaged in super-resolution
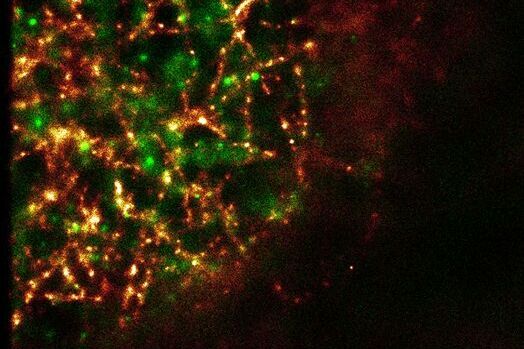
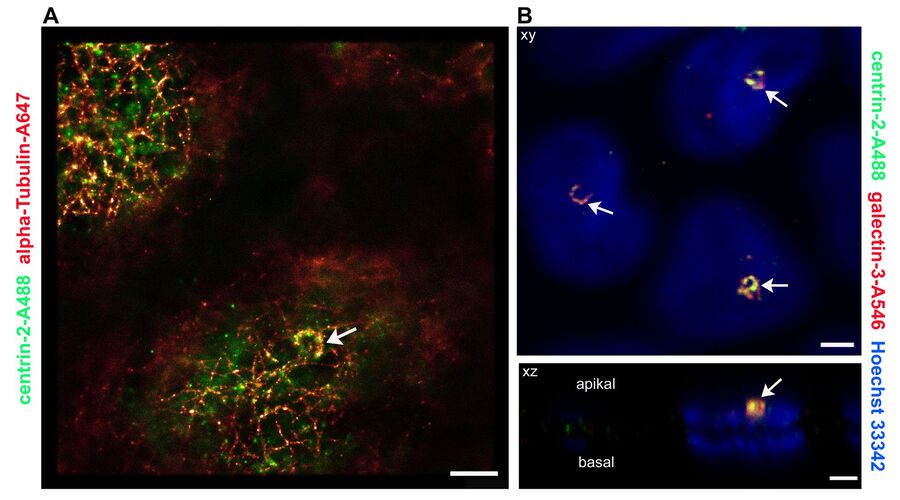
The Leica SR GSD (Leica Microsystems, Wetzlar) is a new multi-purpose microscope that can be used not only for super-resolution GSDIM imaging down to a resolution of 20 nanometers, but also for TIRF microscopy (Total Internal Reflection Fluorescence) and for high-speed widefield live cell imaging. This gives novel and extremely detailed insights into structures of the subapical membrane and their protein composition. Centrin-2 was double-stained with tubulin using an immunohistochemical method and imaged with the GSDIM technique (Figure 2A). The microtubuli network underneath the apical membrane can be clearly visualized. Other visible features are semi-circular to circular central structures that appear once per cell. Arranged around the basal body, these structures are also stained with centrin-2. We were able to see a similar distribution pattern of centrin-2 prior to this as well. Figure 2B shows a double staining of centrin-2 with its binding partner galectin-3 (Koch et al., 2010). However, the conventionally produced images give a much more diffuse view of the protein distribution than the super-resolution GSDIM image.
Fig. 2: Comparison of super-resolution GSDIM microscopy and confocal microscopy. A: For GSDIM microscopy (Leica SR GSD), MDCK cells were cultivated on coverslips, fixed and immunostained with anti-centrin-2/A488 and anti-α tubulin/Alexa647. Tubulin and centrin-2 label a semicircular structure in the basal body area (arrow tip). Scale: 1 µm. B: A confocal microscope (Leica TCS SP2) scan of the apical region of MDCK cells which were immunostained with anti-centrin-2/A488 and anti-galectin-3/Alexa546. The nucleic cells are stained with Hoechst 33342. The x/y image (above) again shows the crescent-shaped centrin-2 structures that also contain the binding partner of centrin-2, galectin-3 (arrow). The x/z image clearly shows that these structures are located apically above the cell nucleus. Scale: 1 µm.
Outlook
The GSDIM technique has engendered a further breakthrough in the light microscopic imaging of cellular structures below the diffraction limit. For sample preparation, the same immunohistochemical methods can be used that are already established in most laboratories for diffraction-limited widefield and confocal microscopy. The GSD microscope can reveal structures that have been invisible up to now. This can be seen by the highly detailed imaging of proteins and their interaction partners, which provides deeper insights into fundamental processes inside the cell. In future, this technology will be useful for gaining new information on the cellular causes of previously incurable diseases.
References
- Bacallao R, Antony C, Dotti C, Karsenti E, Stelzer EH and Simons K: The subcellular organization of Madin-Darby canine kidney cells during the formation of a polarized epithelium. J Cell Biol 109 (1989) 2817–32.
- Bierwagen J, Testa I, Fölling J, Wenzel D, Jakobs S, Eggeling C, Hell SW: Far-field autofluorescence nanoscopy. Nano Lett 10:10 (2010) 4249–52.
- Testa I, Wurm CA, Medda R, Rothermel E, von Middendorf C, Fölling J, Jakobs S, Schönle A, Hell SW and Eggeling C: Multicolor fluorescence nanoscopy in fixed and living cells by exciting conventional fluorophores with a single wavelength. Biophys J 99:8 (2010) 2686-94.
- Fliegauf M, Benzing T, Omran H: When cilia go bad: cilia defects and ciliopathies. Nature Rev Mol Cell Biol 8 (2007) 880–893.
- Fölling J, Bossi M, Bock H, Medda R, Wurm CA, Hein B, Jakobs S, Eggeling C and Hell SW:Fluorescence nanoscopy by ground-state depletion and single-molecule return. Nature Meth 5 (2008) 943–945.
- Koch A, Poirier F, Jacob R and Delacour D: Galectin-3, a Novel Centrosome-associated Protein, Required for Epithelial Morphogenesis. Mol Biol Cell 21 (2010) 219–231.
- Salisbury JL: A mechanistic view on the evolutionary origin for centrin-based control of centriole duplication. J Cell Physiol 213 (2007) 420–428.
- Singla V and Reiter JF: The Primary Cilium as the Cell's Antenna: Signaling at a Sensory Organelle. Science 313 (2006) 629–633.