Unraveling the complexity of TME
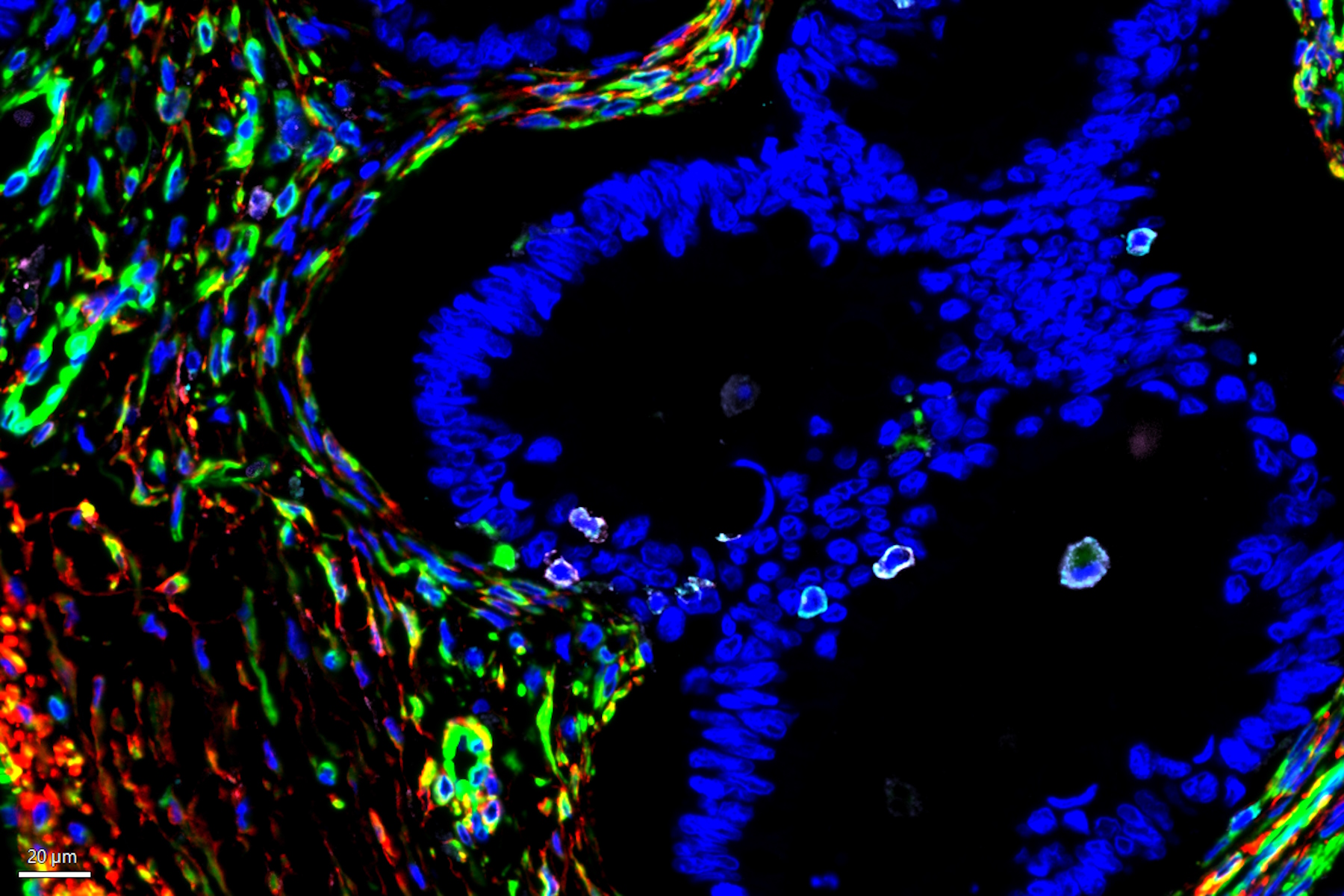
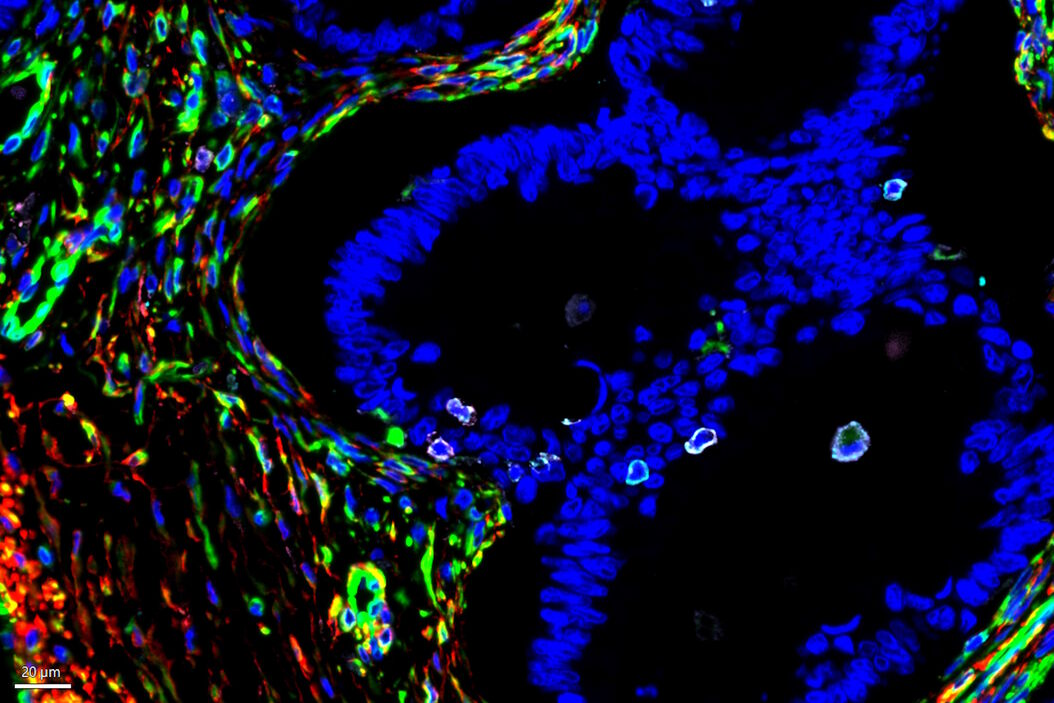
The tumor microenvironment (TME) is heterogeneous and is primarily composed of fibroblasts, extracellular matrix, immune cells, and blood vessels. Importantly, the tumor immune microenvironment (TIME) is a major source of cancer heterogeneity and influences both disease progression and response to therapeutic interventions. Malignant cells typically recruit various cell types such as vascular endothelial cells, cancer-associated fibroblasts (CAFs), and tumor-associated macrophages (TAMs) to promote tumor growth. Therefore, studying such a complex interplay among tumor, stromal cells, and immune cells within the TME necessitates a multiplexed analytical approach to investigate cancerous tissues from diverse origins, to ultimately predict clinical outcomes and design novel therapies. While numerous studies have been focused on investigating the expression patterns of immune-oncology markers within specific tissues, the extent to which such marker expression patterns are shared across tumors originating from various tissues is not adequately understood. In this study, we analyzed a large panel of Cell Signaling Technology (CST®) antibodies that are targeting immune, stromal, epithelial, and vascular markers in cancer tissues derived from various origins including lung, colon, and pancreas. Using multiplexed Cell DIVE imaging, key spatial interactions in the tumor microenvironment that are (1) tissue-specific, and (2) shared across tumors irrespective of origin were determined. Specifically, cluster analysis with Aivia of a common panel of markers across tissues of different origins uncovered molecular signatures that are common to all cancer tissue types as well as those that are unique to specific tissue types to advance our understanding of the disease, and its progression, shedding light on the intricate interactions within the TME.
Open multiplexing solution enables flexibility
Cell DIVE Multiplex Imaging Solution allows probing and imaging of dozens of biomarkers on a whole single tissue section with an iterative staining and dye inactivation workflow. At its core, Cell DIVE is a precise and adaptable open multiplexing solution that enables flexibility in antibody selection of biomarker panels used in a multiplexed imaging study. Cell Signaling Technology (CST) has a broad portfolio of IHC-validated antibodies to detect key proteins in the TME, enabling immune cell detection and phenotyping in tissue. CST offers off the shelf (OTS), ready-to-ship antibody conjugates that have been verified to work on Cell DIVE and offers custom conjugation of IHC-validated antibodies to fluorophores and other detection reagents. CST employs a rigorous approach to IHC validation, followed by verification on the Cell DIVE platform to ensure successful detection of proteins.