Introduction
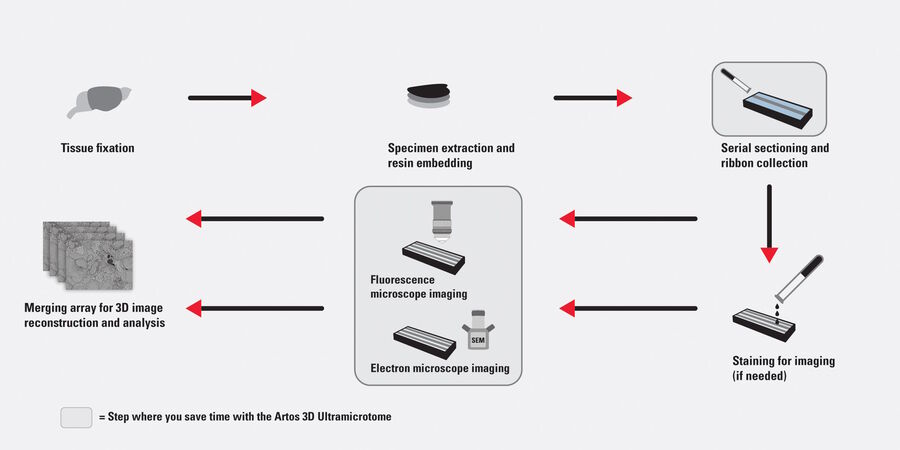
Array tomography (AT) is a high resolution 3D image reconstruction method for cellular and protein structure analysis (refer to figure 1). It exploits the scanning electron microscopy (SEM) imaging of ordered arrays of ultrathin (>20 nm), resin-embedded serial sections of biological specimens collected on silicon substrate carriers [1]. AT allows for quantitative, volumetric structural analysis of many different types of tissues and proteins. As it has better lateral and spatial resolution than conventional confocal microscopy, it can help to better visualize and understand cellular and protein structure. In addition, AT allows cellular and protein structure to be examined in a partially automated manner leading to higher throughput.
Lymph nodes are kidney-shaped organs of the lymphatic system located throughout an animal’s or a person’s body. They are linked by lymphatic vessels and trap foreign bodies from surrounding tissues, helping to initiate the immune response. Lymph nodes are ideal for studying the interactions of immune cells (lymphocytes, dendritic cells, and macrophages), as well as the spread of cancer (tumor metastasis) [2,3]. Precise examination of lymph nodes is critical for these types of immune and cancer studies.
Lymph nodes from mice have been investigated with array tomography using an automated serial sectioning solution.
Serial sectioning with an automated ultramicrotome capable of producing uniform, ultrathin, high quality sections in an aligned array makes the overall workflow for array tomography (3D image data acquisition) more efficient (refer to figure 1). An example of such a serial sectioning solution is the ARTOS 3D ultramicrotome from Leica Microsystems. It automatically produces hundreds of uniform serial sections and avoids wrinkling of the section ribbons during collection due to a specially designed diamond knife. Furthermore, the high section density achieved during collection onto the substrate carrier allows time to be saved during SEM setup. Additionally, AT offers time-saving advantages for specimen preparation when doing correlative light and electron microscopy (CLEM) [4] studies with a transparent section carrier made out of a glass substrate.
Materials and methods
Lymph node isolation
Popliteal lymph nodes were harvested from 6-10 week old C57BL/6 mice. Specimen preparation was done following the OTO (osmium-thiocarbohydrazide-osmium) and ferrocyanide reduced osmium tetroxide fixation protocol of Deerinck et al. [5,6] to enhance the contrast of the tissue for SEM imaging.
The mice were anesthetized with an intraperitoneal injection of 100 mg/kg (wt/wt) ketamin, 10 mg/kg (wt/wt) xylazine, and 3mg/kg (wt/wt) acepromazin. They were then perfused transcardially with phosphate buffered saline (PBS) [0.025 M phosphate buffer, 0.9% (wt/vol) NaCl, pH 7.4] for 2 min. Afterwards, the PBS was replaced via perfusion for 5-10 min with a fixative 0.1 M phosphate buffer (PB) solution having 2% (vol/vol) paraformaldehyde and 2.5% (vol/vol) glutaraldehyde. Immediately after perfusion, the lymph nodes were harvested and post-fixed for another 2h at room temperature (RT).
Specimen preparation
Fixation and contrast enhancement
After harvesting and post-fixation, the lymph node specimens were washed in 0.1 M PB (pH 7.4). Then, to enhance contrast during array tomography (AT), the specimens were immersed for 1 hour in 0.1 M PB with 2% (wt/vol) osmium tetroxide (aqueous) and 1.5% (wt/vol) potassium ferrocyanide on ice and in the dark. Afterwards, they were washed with MilliQ water (5 x 3 min at RT), transferred into filtered thiocarbohydrazide for 20 min at RT, and then again washed with MilliQ water (5 x 3 min at RT). The specimens were placed in 2% (wt/vol) osmium tetroxide (aqueous) for 30 min at RT in the dark and then again washed with MilliQ water (5 x 3 min at RT). They were incubated in 1% (wt/vol) uranyl acetate (aqueous) and placed in the refrigerator at 4°C overnight. The next day, the specimens were washed in MilliQ water (5 x 3 min at RT), incubated in a Walton’s lead aspartate solution at 60°C for 30 min, and washed in MilliQ water (5 x 3 min at RT). They were dehydrated using a graded ethanol series (50%, 70%, 90%, 96%, and 100%) for 5-10 min each and then washed in anhydrous acetone (2 x 10 min at RT).
Resin embedding
The specimens were infiltrated with resin by immersing them in a graded series of DurcupanTM ACM hard resins dissolved in acetone (25%, 50%, and 75%) for 1-2 hours each. Then, they were placed in 100% Durcupan resin overnight and transferred to BEEM capsules filled with fresh resin. The recipe used for the DurcupanTM ACM resin (Sigma-Aldrich) in terms of its 4 components (A-D) was: A) 11.4 g, B) 10 g, C) 0.3 g, and D) 0.1 g. Finally, for polymerization of the resin-embedded specimens, they were kept in an oven at 60°C for 3 days.
Specimen trimming
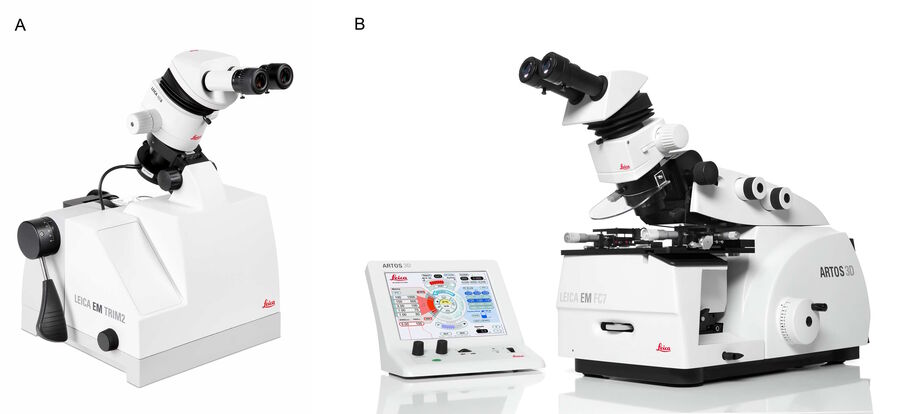
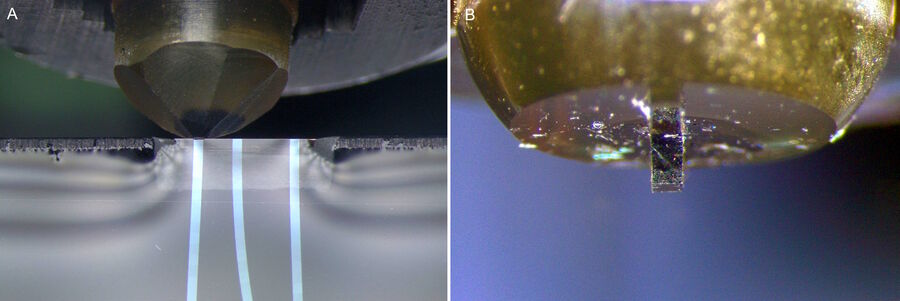
The resin-embedded mouse lymph node specimens were trimmed with the EM TRIM2 trimming device from Leica Microsystems (refer to figure 2A). For straight ribbon formation to occur during the sectioning process, the specimen must have parallel edges. Normally, the specimen is trimmed to this shape with a high-speed milling system, such as the EM TRIM2 or EM Rapid solution.
Specimen sectioning
The trimmed, resin-embedded mouse lymph node specimens were sectioned with the ARTOS 3D ultramicrotome from Leica Microsystems (refer to figure 2B). Additional reasons why an efficient sectioning workflow is possible with the ARTOS 3D solution include:
- Fully motorized movement of the knife stage and auto-transition between section ribbons (figure 3A)
- Library of sectioning protocols available for use with new samples
- Specimen autotrim mode for practical sectioning when preparing large volumes of 3D image data (figure 3B)
- Digital East-West measuring function; and
- Automatic approach of the selected knife segment.
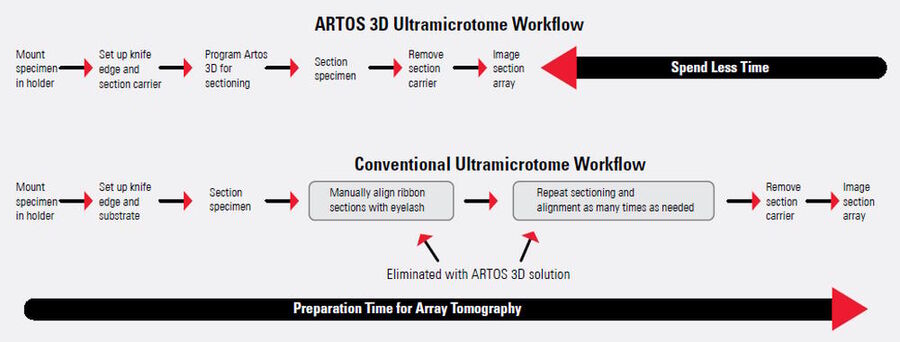
The automation of serial sectioning achieved with the ARTOS 3D solution enables greater workflow efficiency compared to a conventional ultramicrotome. A comparison of the workflow for a conventional ultramicrotome, such as the EM UC7 system, and the ARTOS 3D are shown in figure 4. The work steps of conventional ultramicrotomy which are eliminated with the ARTOS 3D solution are denoted.
Imaging/Array tomography (AT)
After sectioning, the lymph node arrays were transferred to a silicon substrate carrier. Then, AT was performed with SEM to study the targeted proteins in-depth. High resolution images of the lymph node sections were acquired with AT using a field emission SEM (FE-SEM, Merlin compact VP, Zeiss) equipped with the Atlas 5 Array Tomography system. Secondary electron detection was performed at 5 kV.
Image reconstruction
Image reconstructions in 3D were performed as follows:
- Section image dataset was aligned and annotations marking the sub-cellular structures manually made using the TrakEM2 software plugin [7] for the FIJI platform [8] and
- Binary exports of segmented cellular structures were imported into the software Imaris (version 9 Bitplane) [9] for 3D reconstruction and visualization of the iso-surfaces of the T-cells.
Results
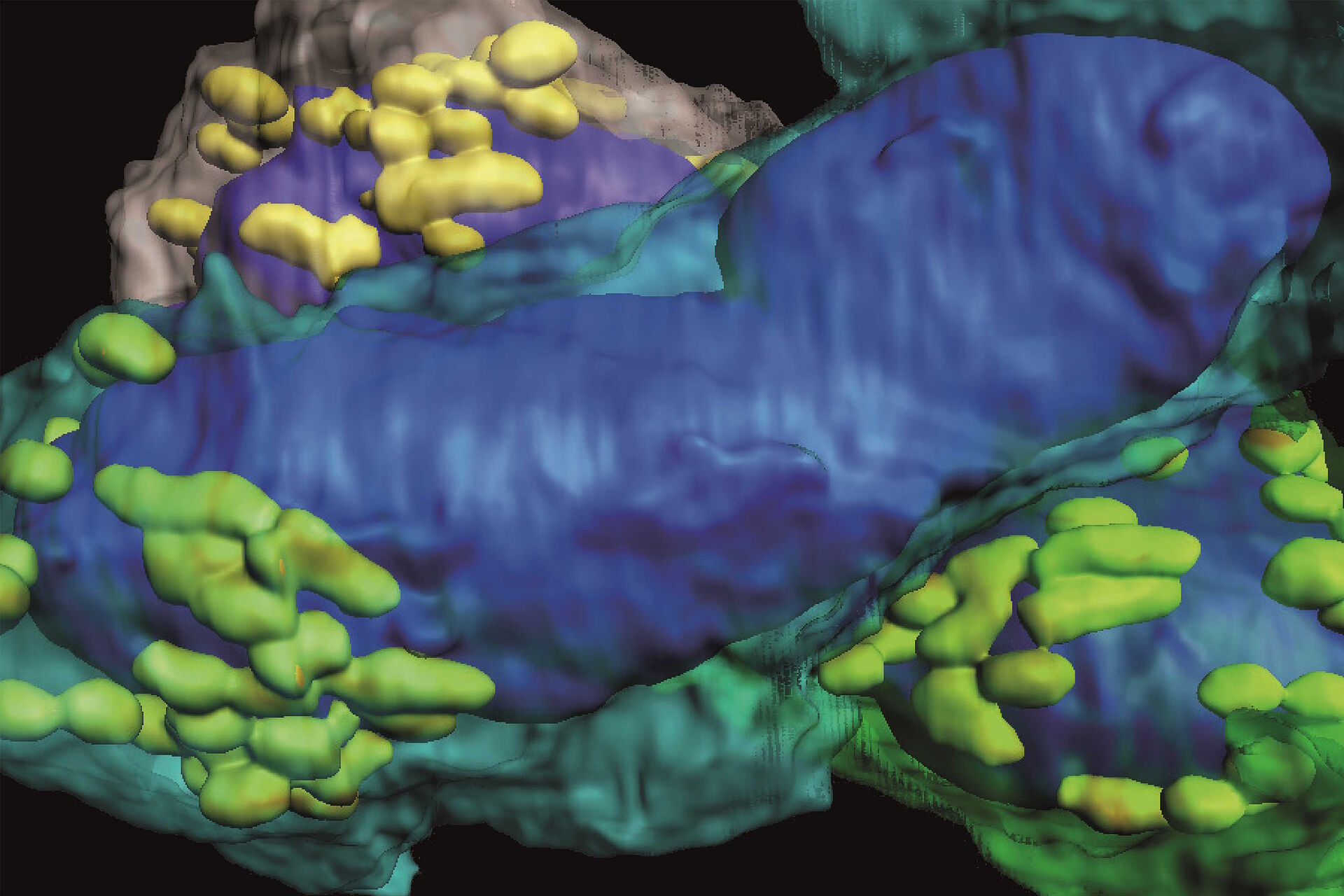
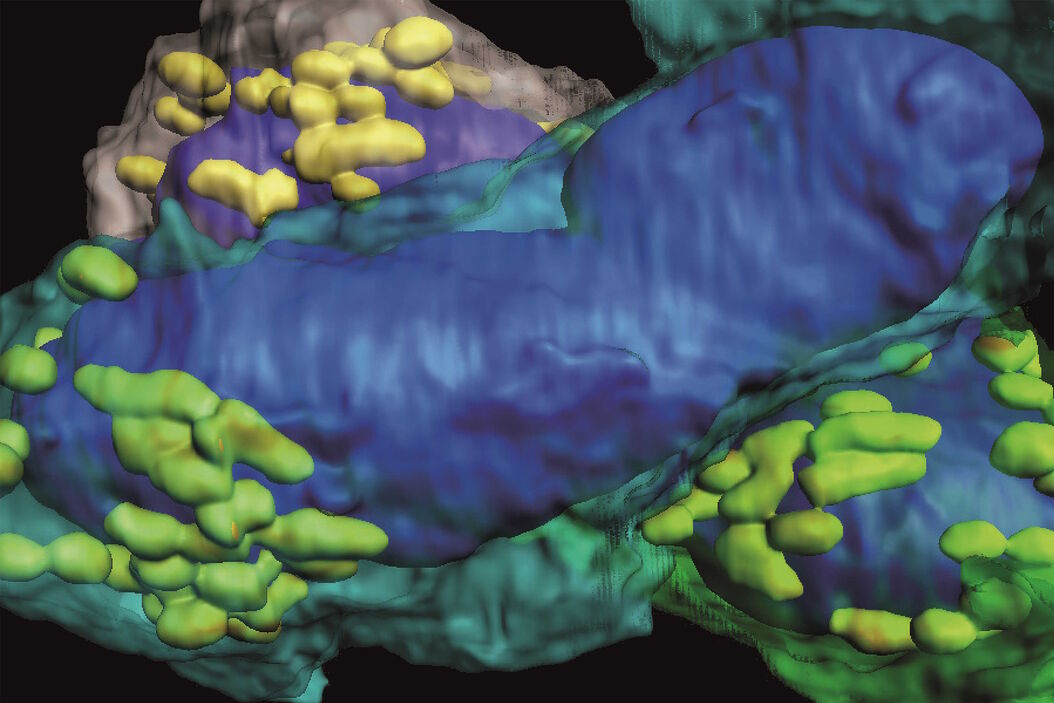
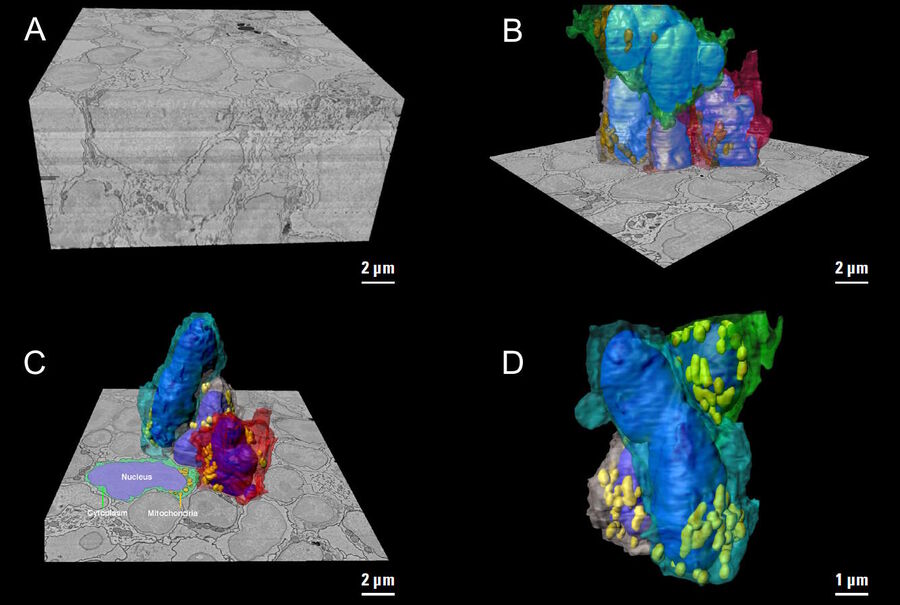
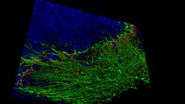
The goal of the mouse lymph node study is to elucidate the complex 3D organization of its paracortical conduit system which transports filtered lymph (containing small soluble antigens) from the periphery towards the venous circulatory system [10]. High resolution 3D image reconstruction of the lymph node ultimately should show how the stromal, myeloid and lymphoid cells spatially interact and have access to the lymph node conduit system.
Three-dimensional (3D) images of the paracortex of a mouse popliteal lymph node were reconstructed using the array topography method (refer to Fig. 5). The images show the 3D volume and shape of the T-cells migrating within the lymph node cells, plus their sub-cellular structures, such as the nucleus, mitochondria, and cytoplasm.
Summary and conclusions
The array tomography method has paved the way for high resolution, reproducible 3D image reconstruction results which allow significant advances in cell biology.
The ARTOS 3D ultramicrotome renders specimen preparation and section alignment for array tomography (AT) more simple leading to a more efficient overall workflow. It achieves this higher efficiency thanks to a specially designed diamond knife, drain tray, and boat for easy collection of wrinkle-free section ribbons on the section carrier. Its wafer section carrier is capable of a high ribbon density and is small enough for optimal loading of multiple carriers into a SEM at the same time. Thus, hundreds of sections can be imaged via SEM without reloading. Transparent materials can also be used for the section carrier, so the ARTOS 3D ultramicrotome is also excellently suited for correlative light and electron microscopy (CLEM) [4].
Acknowledgments
We would like to thank Frank Assen, Ludek Lovicar, Vanessa Zheden, and Michael Sixt of the Institute of Science and Technology (IST) Austria for providing the array tomography image data.