Multi-Color Caspase 3/7 Assays with Mica
From imaging to analysis
Caspases are involved in apoptosis and can be utilized to determine if cells are undergoing this programmed cell death pathway in so-called caspase assays. These assays can be run by e.g. flow cytometry, plate readers, or on a microscope which adds visible, structural information to the quantified data. In this article we describe how Mica can be used to perform caspase 3/7 assays.
With the help of tools like the Navigator or Pixel Classifier, Mica makes it easier to set up, execute and analyze caspase 3/7 assays, even for inexperienced users. Additionally, with the FluoSync technology embedded into Mica, you can image up to 4 fluorophores simultaneously and investigate the fate of additional cellular components during apoptosis with 100% spatiotemporal correlation.
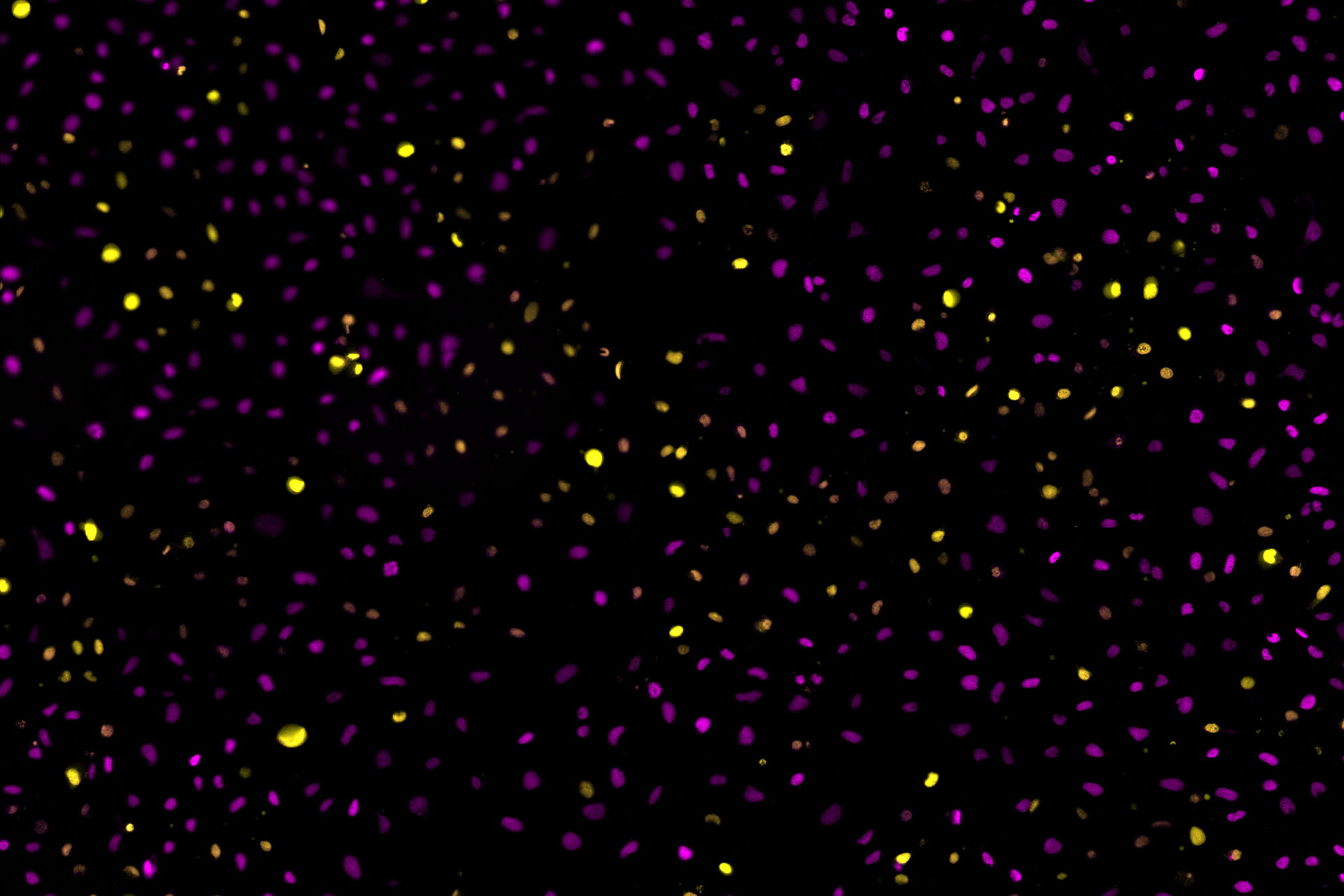
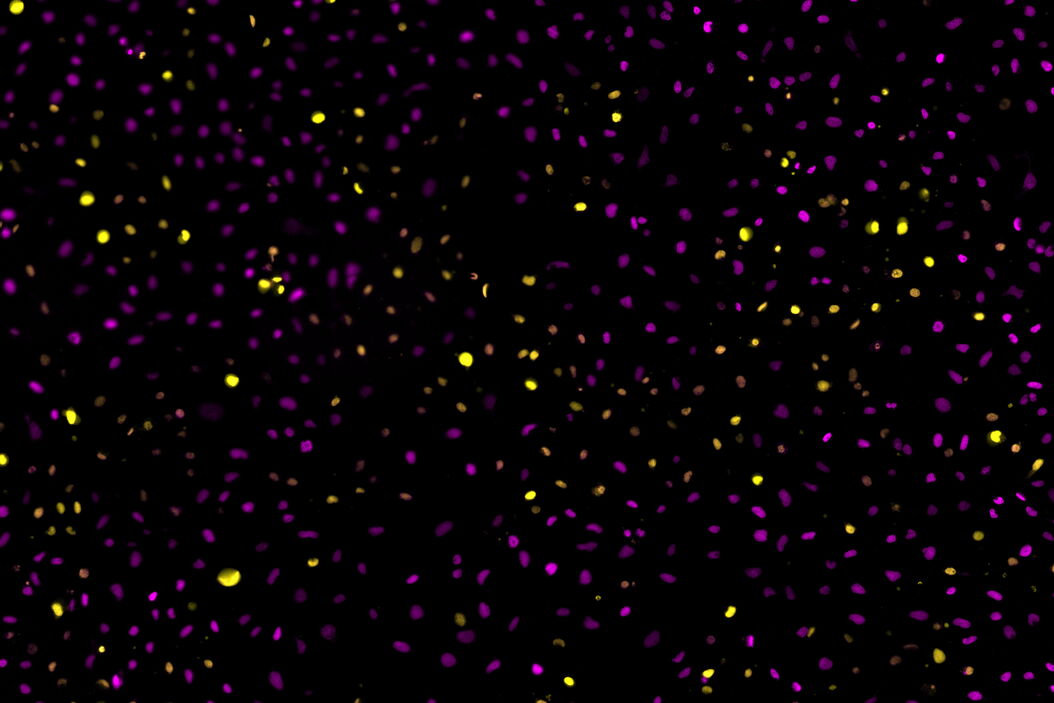
Image: Two-color caspase assay with tile scan. U2OS cells were treated with the nuclear marker DRAQ5 (magenta) and CellEvent™ (yellow). 4 mM staurosporine was added to induce apoptosis. A tile scan of 2x2 FOVs was acquired every 30 minutes for 16 hours duration with the two fluorescence channels using a 20x objective.