Limitations of previous methods
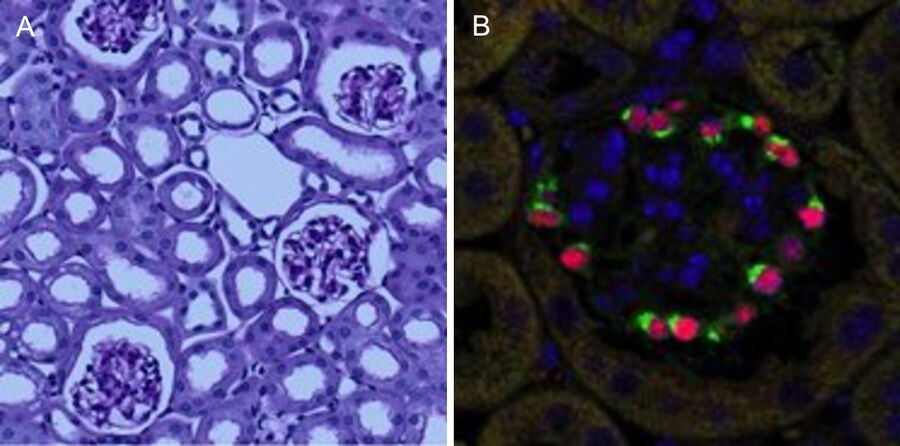
Podocytes are key cellular components of the glomerular filtration barrier in the kidney1. However, they are terminally differentiated and thus incapable of completing successful cell division under normal conditions. As a result, podocyte depletion (reduction in total number per glomerulus or density per glomerulus), is a significant indicator of the development of progressive kidney disease2. Podocyte depletion has, in fact, become a unifying principle of glomerular disease, and to date is routinely assessed by counting podocyte nuclei per glomerular cross-section in histological sections3. Recent studies have, highlighted the limitations of this approach, which include low levels of accuracy and precision, and a high degree of biological variability (particularly in studies in mice).
Stereological methods, both “model-based” and “design-based” (unbiased) methods, produce estimates of podocyte number determined from sampled sections. Model-based methods require knowledge or assumptions of podocyte nuclear size and shape. Generally, these values are assumed, and to the extent these assumptions differ from true values, the estimates obtained are biased. Design-based stereological methods used to estimate podocyte number are extremely time-consuming and have not been adopted by more than a handful of laboratories. Alternative methods for podocyte number estimation using serial histological sections4 (“exhaustive enumeration”) are extremely tedious and laborious.
The combination of tissue clearing and optical sectioning enable new approaches
Fortunately, the advent of new tissue clearing techniques5 in combination with optical sectioning, achievable through confocal, multiphoton or lightsheet microscopy, now make exhaustive enumeration not only attractive but a significant improvement for podocyte quantification, and any other cell of interest, especially those that play central roles in the development and progression of disease.
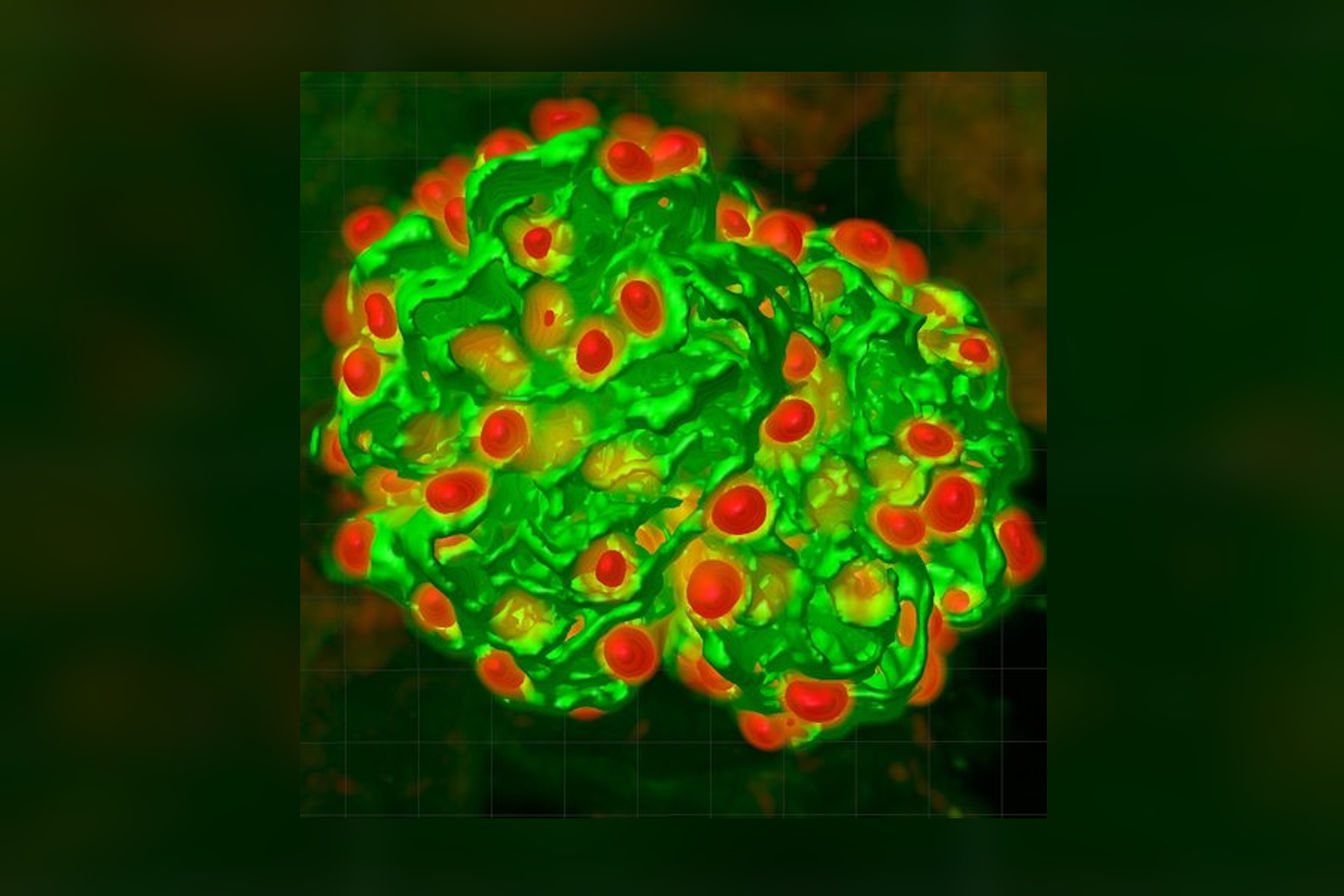
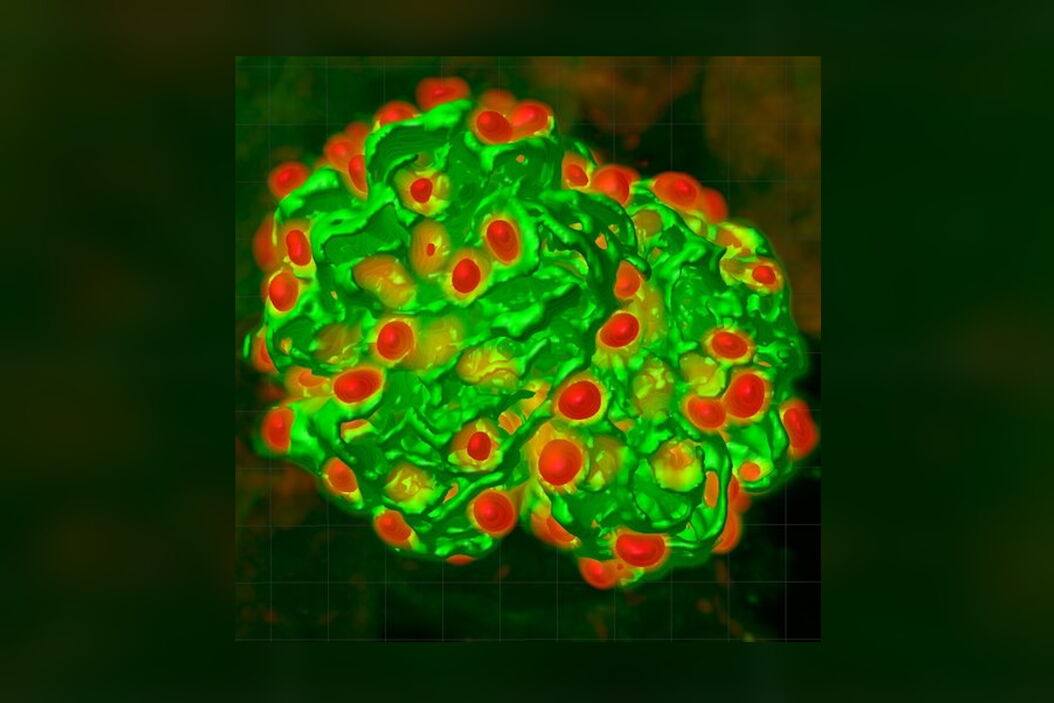
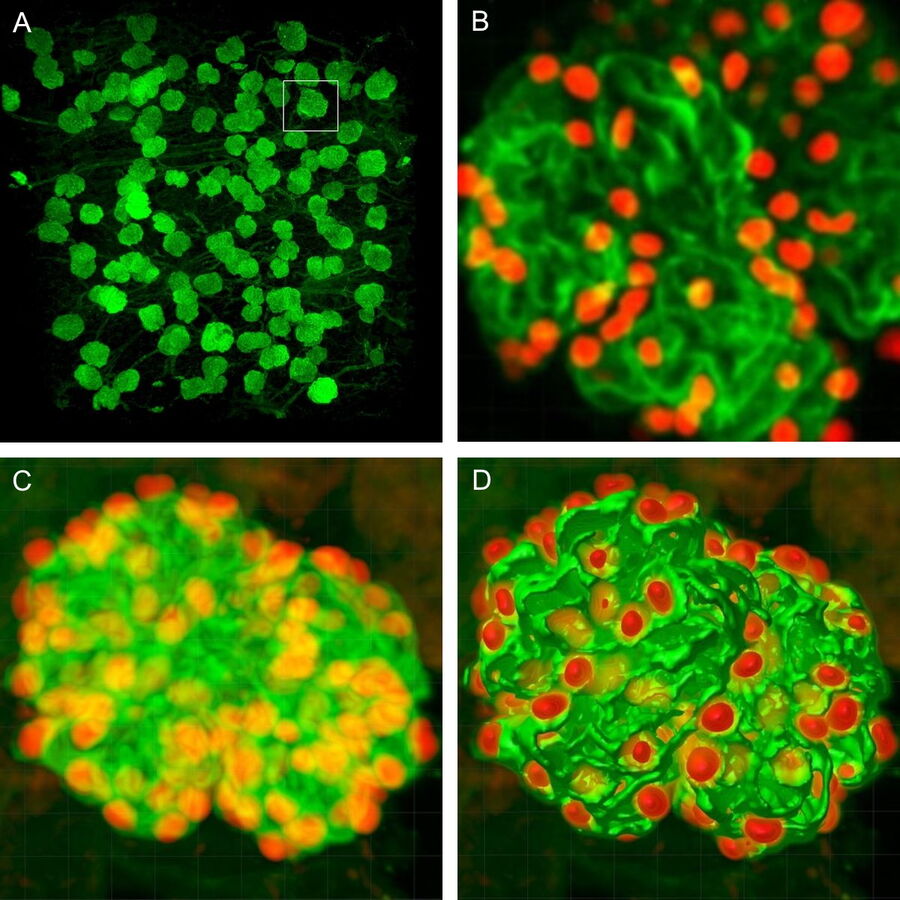
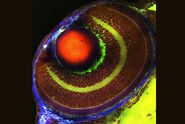
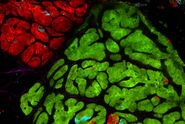
We have adopted this approach, and developed a novel, time-efficient method to study podocyte depletion using a combination of immunofluorescence, optical clearing, confocal microscopy and 3D analysis6. The technique has been used to quantify features of podocyte depletion in transgenic mice expressing the human diphtheria toxin (DT) receptor constitutively in podocytes, which allows conditional and dose-dependent podocyte death.
Our quantitative morphological tool significantly improves the assessment of podocyte depletion, and allows the study of whole glomeruli across the renal cortex. Differences in podocyte depletion between glomeruli in individual kidneys can be detected, a significant advance in the study of focal glomerular diseases.
Development of a novel method for imaging and quantifying podocyte depletion
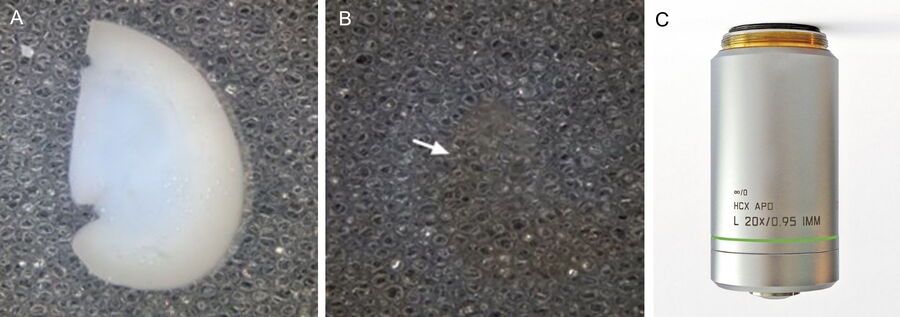
Formalin-fixed kidneys are sliced to 800µm thickness, and the slices then subjected to 1hr antigen retrieval using a modified immunofluorescence protocol. Optical clearing is achieved by embedding the slices in agarose which are then dehydrated through changes in methanol, and cleared by several changes of benzyl alcohol, benzyl benzoate (BABB).
By gluing the edges of the dehydrated agarose to a glass petri dish, we immobilise the thick kidney slices on the bottom of the dish, making the specimens amenable to imaging on an upright Leica TCS SP8 MP confocal/multiphoton microscope equipped with the specialised Leica 20x/0.95 NA BABB immersion objective. The BABB mixture used has a refractive index of 1.56, which also permits higher resolution imaging using x40 and x63 oil objectives to be carried out on an inverted microscope when required.
For podocyte enumeration, 800µm thick kidney slices were imaged with stage-tiling in order to access the full cross-section of the kidney. This facilitated systematic imaging across the renal cortex from superficial to juxtamedullary regions to produce unbiased results. Slices up to 2mm thick (which represents the full working distance of this objective) were also imaged, but in general the use of 800µm sections, imaged across 2-3 channels at 1µm intervals provided ideal resolution, working depth and overall imaging time.
The final datasets provided quantifiable podocyte microstructure for analysis, as well as full 3D glomerular visualisation. Using markers for all nuclei (DAPI), podocyte cytoplasm (synaptopodin, SNP) and podocyte nuclei (p57), and the masking and counting features in Imaris (Bitplane AG, Zürich, Switzerland) it was possible to determine for each glomerulus analysed: (i) total podocyte number (ii) mean podocyte volume, (iii) individual glomerulus volume; and from those data calculate (iv) podocyte density (number per volume of glomerulus); (v) mean podocyte nuclear volume; and (vi) mean podocyte cytoplasmic volume.
While the image analysis component was not fully automated and required manual supervision, counting and analysis was significantly better and faster than all other methods thus far. Once confocal images were obtained, values for the above parameters were obtained in approximately 5mins per glomerulus.
Discussion and outlook
Advances in specimen preparation techniques and innovations in imaging continue to push the boundaries of discovery in the life sciences. Our understanding of tissue microstructure and cell and developmental biology has advanced significantly through innovative labelling approaches such as perfusion labelling, improvements in specificity and photostability of fluorescent labels, and tissue clearing techniques.
We now have unprecedented ability to study whole organisms, whole organs and tissues at the cellular and subcellular level. These techniques are applicable to fluorescence widefield, lightsheet and confocal microscopy modalities, all of which are supported by a wide range of high NA, long working distance objectives that match the refractive indices of BABB, Clarity and glycerol.
Moreover, image analysis can be used to efficiently measure number, length, surface area and volume of features of interest. A current challenge arising for many, however, is the handling, analysis and visualisation of these subsequently larger datasets which have now rapidly grown from GB to TB sizes. It is becoming increasingly important to develop better solutions for the rapidly developing discipline of bioimage informatics.
References
- Reiser J, Sever S. Podocyte biology and pathogenesis of kidney disease. Annu Rev Med 2013; 64: 357-66.
- Fukuda A, Chowdhury MA, Venkatareddy MP, et al. Growth-dependent podocyte failure causes glomerulosclerosis. J Am Soc Nephrol 2012; 23(8): 1351-63.
- Puelles VG, Bertram JF. Counting glomeruli and podocytes: rationale and methodologies. Curr Opin Nephrol Hypertens. 2015 May; 24(3):224-30.
- Lemley KV, Bertram JF, Nicholas SB, White K. Estimation of glomerular podocyte number: a selection of valid methods. J Am Soc Nephrol 2013; 24(8): 1193-202.
- Silvestri L, Costantini I, Sacconi L, Pavone FS. Clearing of fixed tissue: a review from a microscopist’s perspective. Journal of biomedical optics. 2016; 21(8):081205.
- Puelles VG, van der Wolde JW, Schulze KE, Short KM, Wong MN, Bensley JG, Cullen-McEwen LA, Caruana G, Hokke SN, Li J, Firth SD. Validation of a Three-Dimensional Method for Counting and Sizing Podocytes in Whole Glomeruli. J Am Soc Nephrol 2016 Mar 14. (published ahead of print, doi:10.1681/ASN.2015121340)