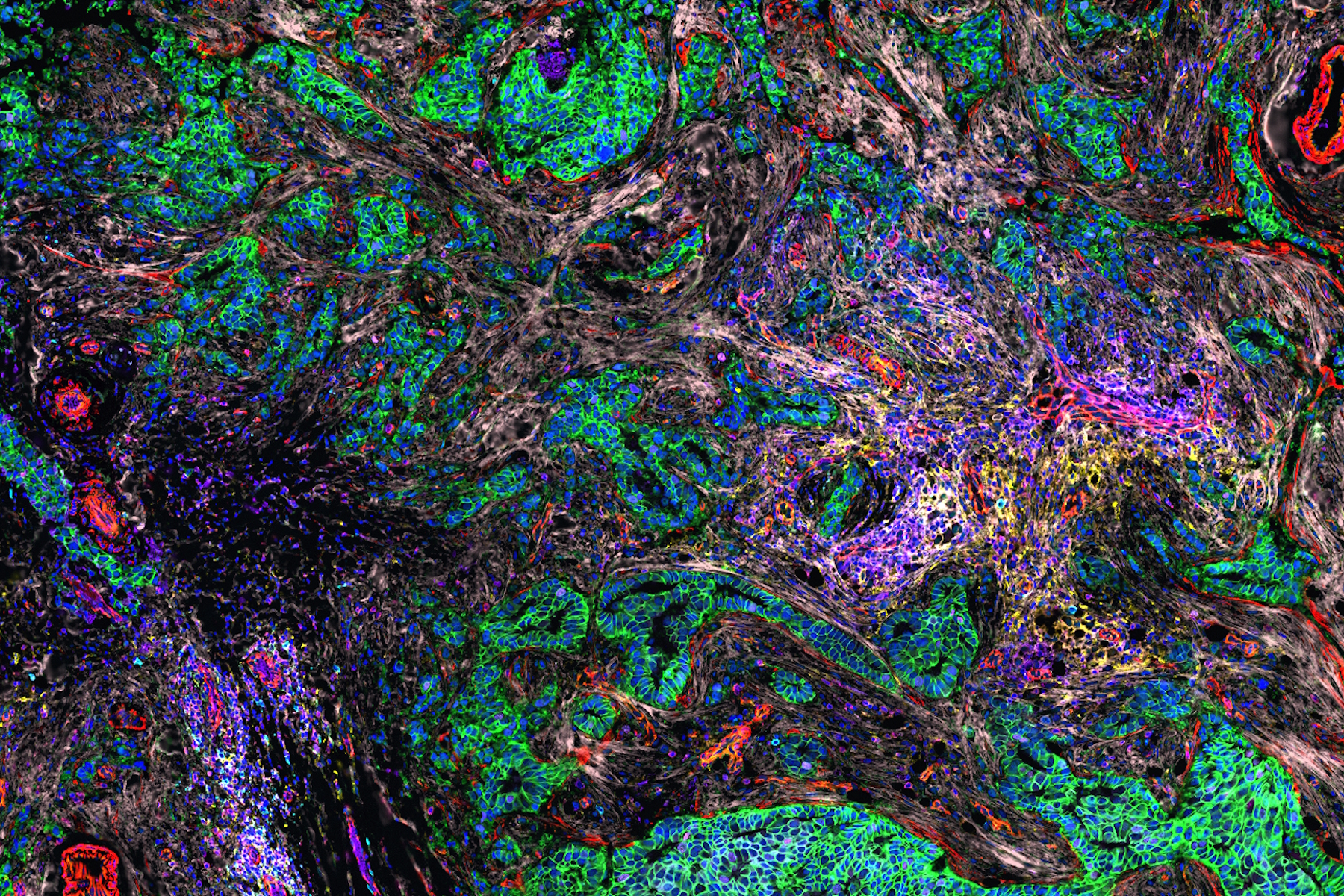
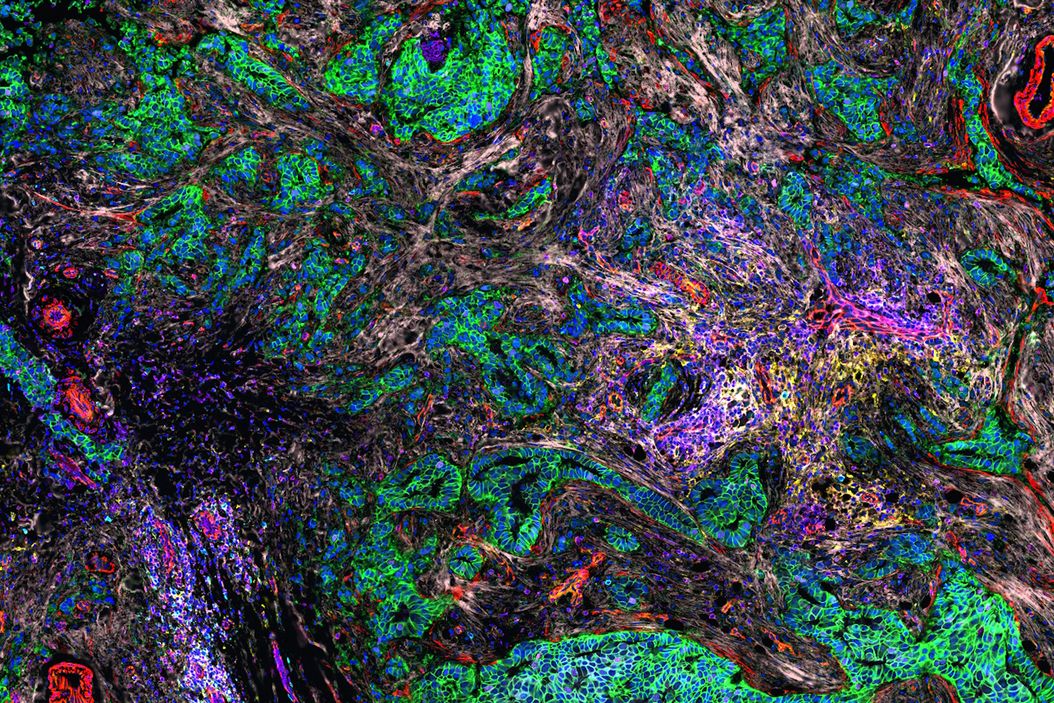
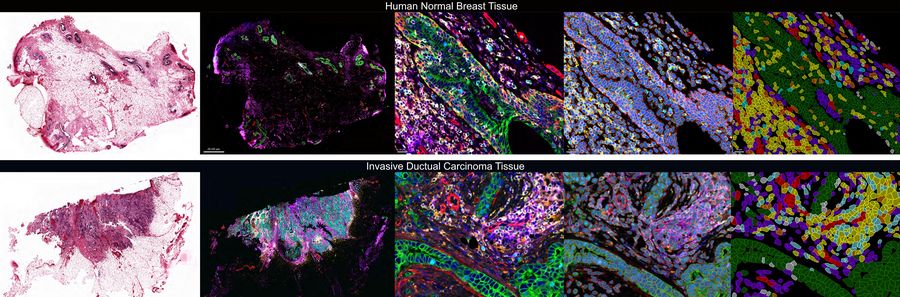
Investigating the TME in breast cancer using multiplexed imaging
Breast cancer (BC) is a heterogeneous disease both at the molecular and clinical level. Worldwide, BC is the leading cause of cancer-related deaths in women. Besides conventional chemo/radio therapy, immunotherapy is one of the most promising choices for a small subset of responding individuals. However, these treatments often fail to prevent recurrence and metastasis. Disease heterogeneity in BC poses challenges for survival prediction, as patients with similar diagnoses often respond differently to treatment. Herein, we apply multiplexed spatial proteomics analysis on breast cancer tissue paired with normal tissue and observe multiple immune cell types and biomarkers that could provide insight into features of the TME that influence response to immunotherapy.
The Cell DIVE Multiplex Imaging Solution allows probing and imaging of dozens of biomarkers on a whole single tissue section with an iterative staining and dye inactivation workflow. Cell DIVE is an open multiplexing solution that enables flexibility in antibody selection of biomarker panels used in a multiplexed imaging study. Cell Signaling Technology (CST) has a broad portfolio of IHC-validated antibodies to detect key proteins in the TME, enabling immune cell detection and phenotyping in tissue. CST offers off the shelf (OTS), ready-to-ship antibody conjugates that have been verified to work on Cell DIVE. CST employs a rigorous approach to IHC validation, followed by verification on the Cell DIVE platform to ensure successful detection of proteins. Here, we demonstrate multiplexed Cell DIVE imaging using a novel panel of dozens of CST biomarkers across human breast cancer tissue samples.
Unveiling the tumor-immune landscape using AI-powered spatial analysis
AI-guided analysis using Aivia software was utilized to analyze the human normal breast and invasive ductal carcinoma (IDC) tissues which revealed single cells characterized by expression of 40+ antibodies targeted towards various phenotypes (Fig.1). A comparative analysis of the tissues revealed alterations in tumor growth, immune evasion, metabolic, stromal, and stem-cell associated markers, and immune cell subtypes such as regulatory T cells (T-regs), cells expressing immune checkpoint proteins, B cells, macrophages, and dendritic cells. Clustering analysis demonstrated differences in co-expression profiles of cell phenotypes in IDC and control tissues. We also identified diverse spatial mapping of cell types in relation to the tumor for local neighborhoods indicating a heterogenous immune landscape (mix of anti and pro-tumor cell types) surrounding tumor cells.
Overall, this study revealed crucial insights about the TME, including differences in global expression levels, co-expression patterns, spatial positioning for control and breast cancer tissues. This information is crucial for understanding tumor behavior, predicting outcomes, and designing effective treatment strategies. In summary, multiplexed whole slide imaging using validated antibodies and Aivia, AI-based image processing software, enables researchers to gain comprehensive insights into the TME.