Live-Cell Imaging
Live-cell imaging reveals information of biological processes at a microscopic level. It requires that the specimen is kept under near physiological conditions. Some of them are mentioned in the paragraph below. Typical specimens are classical cell cultures, live tissues, organoids, or model organisms. Commonly the sample is investigated with fluorescence microscopy. To achieve this, cells are transfected and, consequently, transgenic organisms are produced which express fluorescently labelled proteins. In addition, live-cell dyes can be used.
Challenges
Environment Conditions:
Live-cell imaging conditions must mimic a physiological environment to achieve as realistic experimental results as possible. Optimal physiological conditions – meaning temperature, pH, oxygen level – differ between organisms. Whereas mammalian cells require ~37°C, insect cells grow best at ~27°C, fish prefer 28°C, while fission yeast is kept at ~30°C.
The pH within cell culture media is kept constantly at 7.0 – 7.4 with the help of a sodium bicarbonate buffer, which requires the presence of CO2 in the incubator atmosphere accordingly. In addition, the incubator atmosphere should be saturated with water, so that no culture medium can evaporate.
In vivo oxygen levels vary across tissues and cell types. Interestingly, currently oxygenation is not widely considered for cell culture incubators and live-cell imaging. Sub-optimal oxygen levels can result amongst others in decreased proliferation rates (hyperoxia) or reduced metabolic rates (hypoxia) (Hadanny, A.; Efrati, S.).
Light protection is another challenge for live-cell imaging, because light exposure can influence living cells per se, as well as the fluorophore. For example, high light doses can lead to DNA damage or photobleach the fluorophores.
Mica has been designed to cope with all these challenges:
The housing itself acts as an incubator. This means that the whole volume around the sample is heated up to the required temperature (5°C above room temperature up to 42°C) and equilibrated to the desired CO2 level and humidity.
The O2 levels are controlled within an additional stage-top chamber, if desired. All parameters can be controlled by the Mica LAS X software. Light exposure of the sample is set by the OneTouch function and can be tuned with the “Sample Protection – Image Quality” slider to balance the required amount of signal vs. protecting the living cells and fluorophores.
For sample access with minimal environment disturbance, there is a small service flap hidden in the front door.
Light can be toxic
As light is energy that you bring into the sample, it can have negative effects. Not only UV light, which, for example, can be harmful to the skin, any light might interfere with molecules within the cell. Additionally, fluorophores may form radicals that interact with molecules. The negative effects of light are known as phototoxicity. The challenge is to balance the amount of light needed to yield enough signal to answer the scientific question and minimize phototoxicity. Mica helps users here by offering a tool to balance exactly these 2 things without users having to know the technical background – the protection - quality slider. With a single click, OneTouch will set up all excitation and detection parameters according to the selection made on the slider.
Intracellular dynamics can be very fast. For example, molecular motors which carry organelles or vesicles reach a speed up to several µm/s (Alberts B. et al.). This influences the requirements for the image acquisition, especially when more than one fluorophore should be imaged. The problem here is that biology does not pause for all fluorescent channels so that it can be acquired in the identical state, but progresses continuously. In a classical fluorescence filter-based system, commonly the most time-consuming step is to change the filter cubes for the different channels. This leads to the fact that two fluorophores are monitored at two different time-points in the experiment, making exact spatiotemporal correlation impossible.
The Mica FluoSync™ technology enables users to acquire up to four fluorescence channels simultaneously. This means that different vesicle cargoes, cytoskeleton, and motor proteins can be imaged with 100% spatiotemporal correlation. In addition, simultaneous acquisition enables faster cadence of imaging time-points, thus, fast cellular movements can be recorded with higher time-resolution.
Recording many repetitions with a given time frame
It can be a challenge to record many repetitions within one experiment. Assuming that a user wants to record every 10 minutes, there are only so many positions that can be recorded before the next cycle begins. Normally, this limitation becomes exacerbated if you want to record multiple labels in the same position, but this is not so with Mica. The FluoSync™ technology enables users to record up to four times faster, as it acquires up to four labels simultaneously, thus leaving more time for users to record more positions. Users do not have to trade off the number of labels and positions anymore.
Finding the right positions to image
Finding and setting up the experiment at the right position on the sample can be challenging. Using eyepieces, one has to maintain a mental map of the sample and individual positions have to be remembered. Using a digital microscope to generate an overview of the sample may help, but one still needs to be able to point to a position in the image which is intended for further imaging. Mica offers a way to make this simple using the Navigator tool. Overviews of the sample at lower or higher magnification help users locate the interesting positions. These regions of interest can be directly marked by drawing on the images with the tool. That way the context of the subsequent high-resolution imaging can be kept.
Choosing the right objective for live-cell imaging
As live-cell imaging is typically done in aqueous solutions, high magnification objectives are designed to use water as an immersion medium, because it matches the optical properties of the media. Placing water between the objective and sample is cumbersome and can lead to changes of the focus and position of the sample. Furthermore, water can evaporate rather quickly, thus it needs constant replenishment. Mica has integrated a feedback loop to first establish and second maintain the immersion throughout the usage of the water immersion objective. This approach leaves no manual interaction to be done and allows long-term experiments.
To further increase the optical quality, some objectives feature a CORRring to compensate for the thickness of the sample carrier. The CORRring can require either manual or motorized operation. Due to the SmartCORR feature, the optimization is automatically done with Mica.
Comparability between repetitions
One key aspect of scientific experiments is to change as few variables as possible to pinpoint any effects on the sample and results. This includes applying the same set of imaging conditions throughout all experiments. One aspect is stable environmental conditions (temp, pH, and O2, see also above). The next important aspect is the same imaging parameters (excitation and detection). Mica by default keeps the imaging parameters constant within projects and only changes them at the will of users. Imaging parameters can be restored from reference images. In terms of the environmental conditions, Mica is an incubator. Conditions are stable to allow the growth of MDCK cysts for 3 consecutive weeks (see also below).
Methods
Mica has special specimen holders for live-cell imaging. This includes holders for small (36 mm) and medium (60 mm) size Petri dishes.
Vesicles:
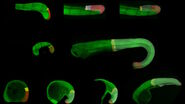
U2OS cells were simultaneously stained with WGA-Alexa488 and WGA-Alexa555. Both dyes accumulate at the plasma membrane and all vesicles and endosomes of the cell. Fluorophores were imaged either simultaneously or sequentially for comparison. The environment was set to keep a constant 37 °C and 5% CO2, thus physiological pH levels.
For high magnification imaging the 63x/1.20 WATER objective with Auto-Immersion was used. No manual interaction was needed.
Zebrafish:
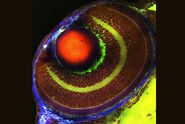
The imaged fish line (Danio rerio) is a transgenic line for a mesodermal fate reporter (Tbxta:GFP). For this experiment a developing embryo was imaged from sphere stage to somite stages. The interstitial fluid was labelled with Dextran-Alexa647. The embryo was kept at 28°C.
Cysts:
a) MDCK cells stably transfected with EB1-GFP were grown in Matrigel to induce cyst growth on ibidi µ-Slide 8 Well slides. The environment was set to keep a constant 37 °C, 5% CO2 and high humidity.
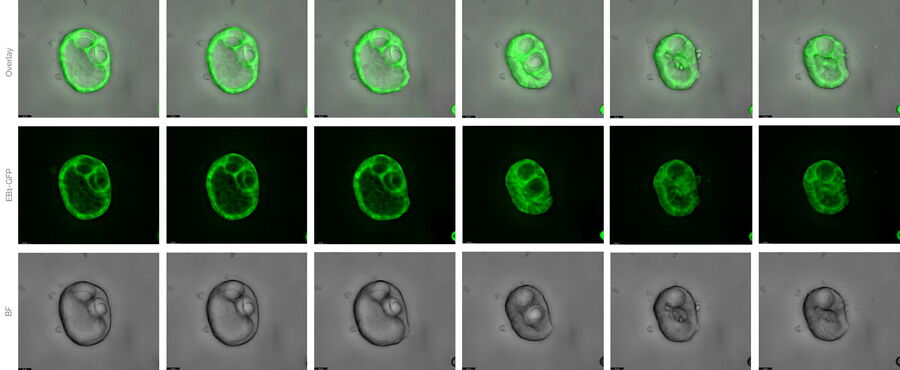
At day 2, the cells showed already 3D conglomerations and were highly mobile inside the Matrigel.
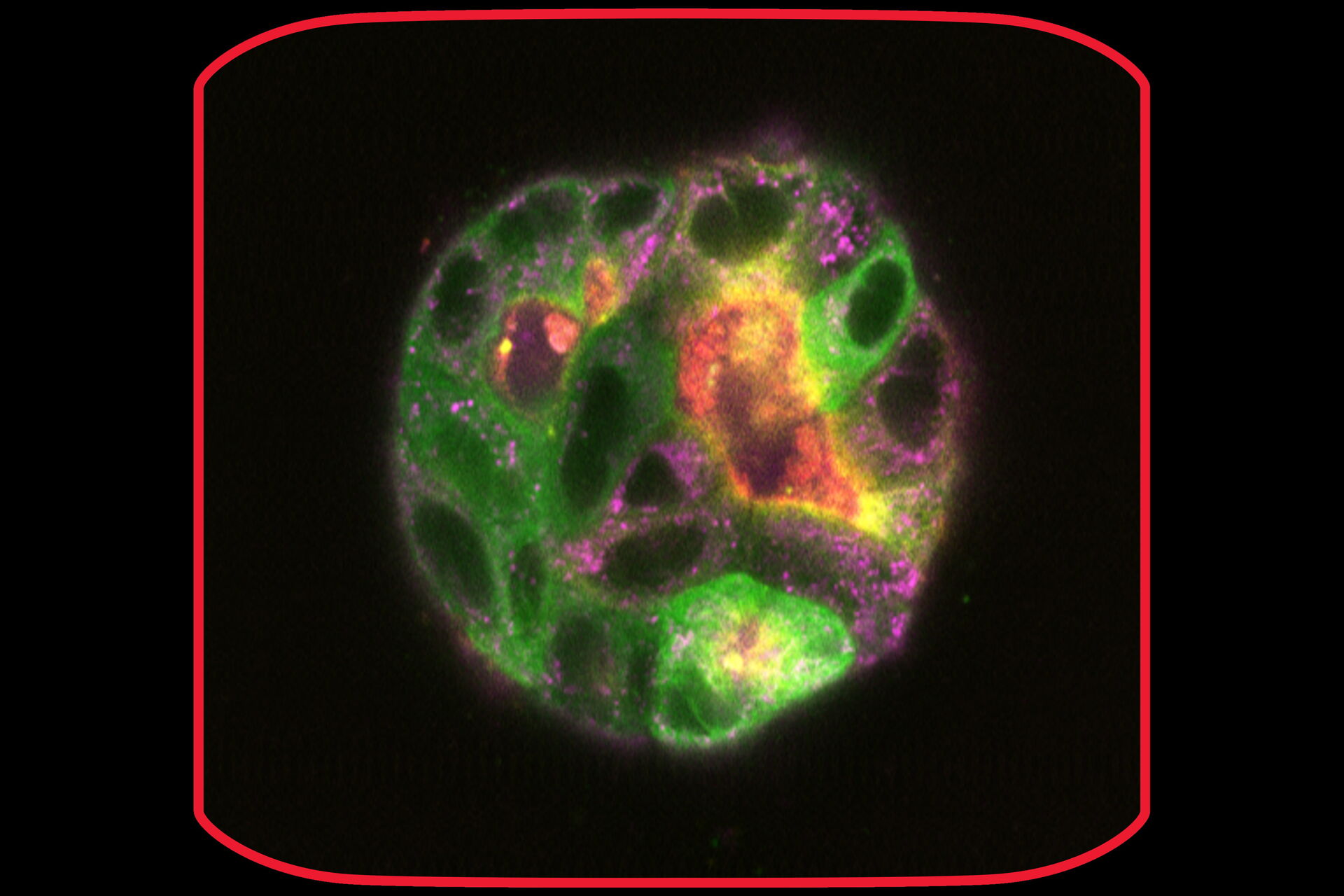
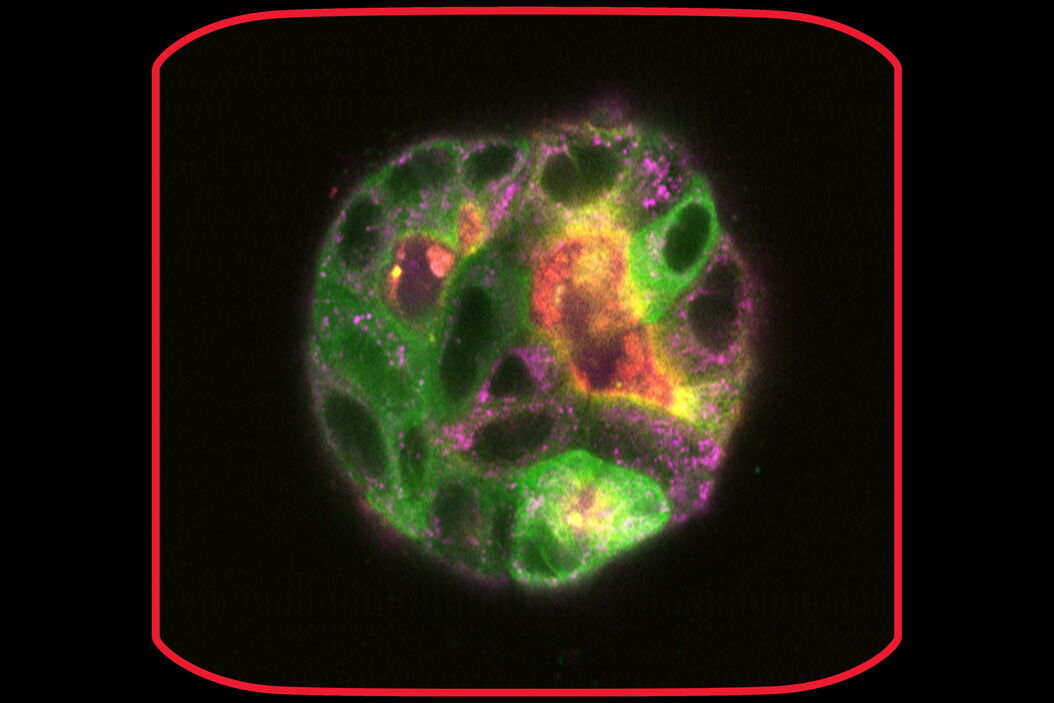
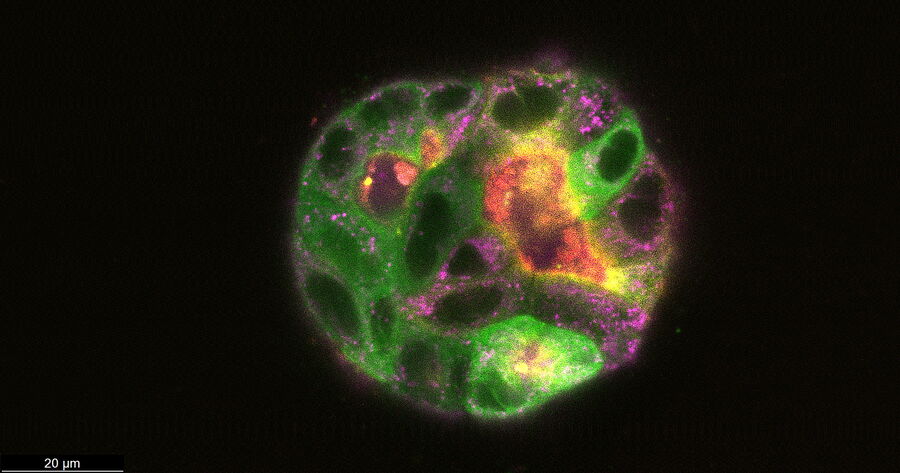
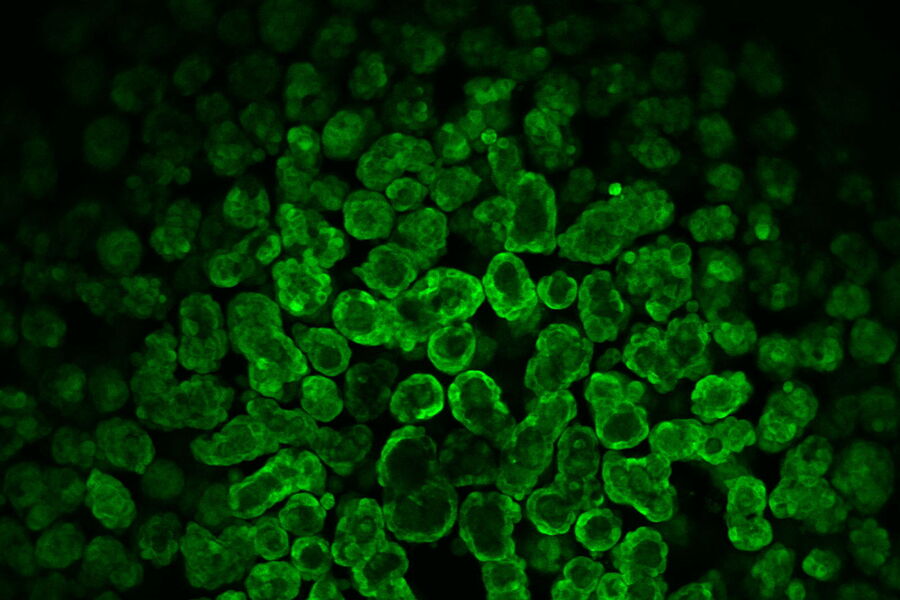
At day 9 the cysts stayed relatively stable in place and were imaged at higher magnification (63x) at steady state and over time (6 days and a 6-hour time-interval in GFP and IMC (a modulation contrast for brightfield imaging). 3D acquisition allowed to get a 3D impression of the cysts (418 layers with 220.7 nm spacing, total Z-Stack size 92.02 µm). At day 9 some of the cysts were labelled with Mito-Blue, SiR-Actin, and Tubulin-SPY555, in addition to EB1-GFP.
b) 5000 MDCK cells stably transfected with EB1-GFP were grown in U-well plates. Interestingly, these cells also formed cyst-like structures after a few days. One of these cyst-like structures was further investigated in confocal mode for 3D reconstruction.
FUCCI - Fluorescence Ubiquitin Cell Cycle Indicator
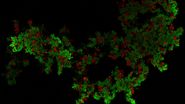
Confluent and quiescent human keratinocytes HaCaT expressing FUCCI construct (GFP-RFP) were imaged to analyze collective cell motility induced by EGF treatment. A FUCCI construct is used as an indicator of the cell cycle phase during the streaming. In each well of a 12-well plate a tile scan of 6 positions was acquired using the 10x objective to capture a large field of view. The total duration was 16 h and 40 minutes with 5 minutes interval.
Results
Short-term experiment with fast events
U2OS cells were labelled with two different fluorescence stains marking the same structure, WGA-Alexa488 and WGA-Alexa555 which both bind to cellular membranes. By acquiring both channels in Mica sequential mode, this experimental setup enables to introduce an artificial time shift between the acquisition of WGA-Alexa488 (green) and WGA-Alexa555 (red). With it, the same membrane structures, which are marked with the two different Alexa dyes are imaged at two minimally shifted timepoints. In the comparison of both methods, you can see that while in the simultaneous recording all vesicles are yellow – a mixture of green and red – in the sequential recording some fast moving vesicles appear to be two different structures – one green and one red vesicle.
Medium-term experiment
The zebrafish is a model organism which is used very often for developmental studies. In this example, the interest was to study the coordination of cells in space which is influenced by the viscosity of the surrounding tissue. Two channels were imaged for 24 hours in THUNDER mode with Large Volume Computational Clearing (LVCC) to get more details.
Mica allowed the simultaneous imaging of GFP and RFP in widefield microscopy. The possibility to work with a filter free system speeds up the acquisition, avoiding the filter shift in between one channel and the other.
The video shows standard widefield image vs. THUNDER Instant Computational Clearing of a representative tile highlighting the increased contrast.
Long-term experiments
MDCK cells are polarized epithelial cells which mature into so called cysts, if cultured on a matrix such as Matrigel. These three-dimensional structures can be used to study developmental processes and the underlying protein transport mechanisms. Here, the cells were stably transfected with a microtubule binding protein linked to GFP and incubated on Mica for a total of 3 weeks.
After 9 days of development, some of the cysts mentioned above were stained with live-cell dyes to visualize actin, tubulin, and mitochondria, in addition to the GFP-tagged microtubule binding protein. Subsequently, cysts were imaged in confocal mode.
Cyst-like structures can also be formed by cultivating cells in U-well plates. In this experiment, MDCK cells were cultured and recorded on Mica from day 1 in brightfield and widefield fluorescence to study cyst formation. The Mica incubator properties allowed the cells to be kept happy for days as they developed into cyst-like structures.
After 14 days of cultivation, the cysts were imaged in confocal mode at low magnification to identify an interesting single cyst. Screening the sample – in this case a 96-well plate – for the right place for further investigation can be cumbersome. The Navigator tool helped here to keep the overview of the sample and easily navigate to relevant regions of interest.
One of the cysts was then imaged in more detail with the 63x objective in confocal mode with LIGHTNING. The immersion for the 63x/1.20 water objective was established automatically and optimized for the sample with SmartCORR. Video 5 shows the comparison of the volume acquired in confocal and processed with LIGHTNING.
Conclusions
Mica is highly adapted for live-cell imaging which helps users get reliable results ranging from fast, short-term experiments up to long-term ones taking place for weeks: The integrated incubation system delivers physiological-like conditions concerning temperature and pH, enabling live-cell imaging for weeks. Also, Oxygen levels can be controlled to achieve higher significant data. The FluoSync™ technology enables the imaging of up to four fluorophores simultaneously which means that fast cellular events can be imaged with absolute spatiotemporal correlation. Moreover, FluoSync™ decreases intervals between recorded multi-color images and, thus, improves temporal resolution. THUNDER and LIGHTNING help users extract more details from the sample, while automated processes like the OneTouch for illumination and automated water immersion give access to all users even with minimal training and limited technical background.