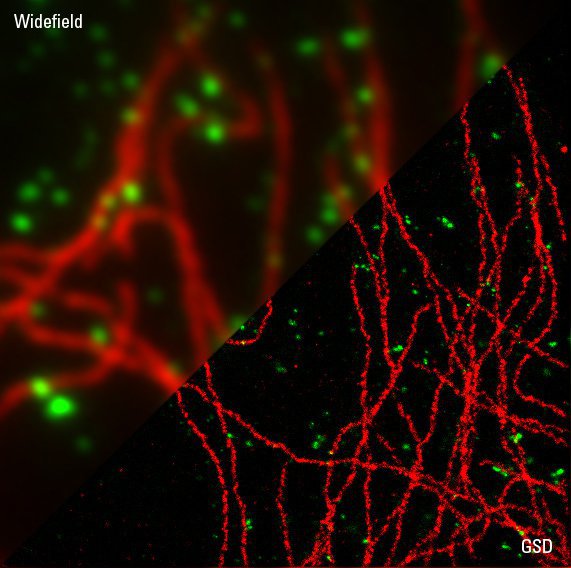
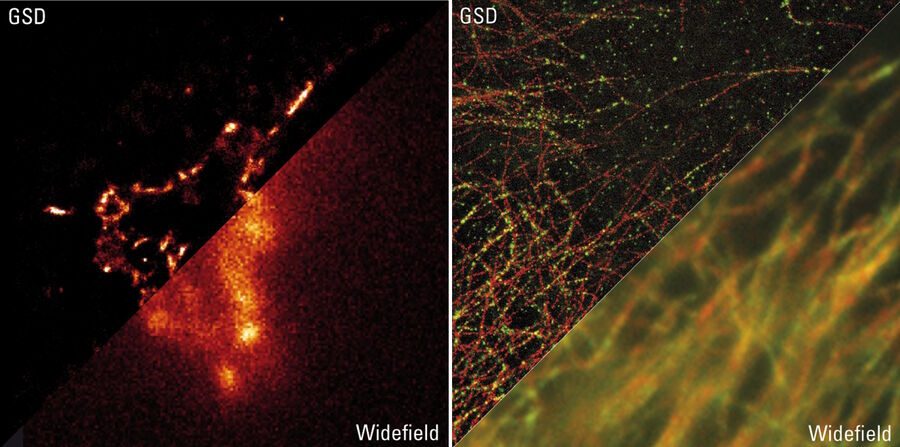
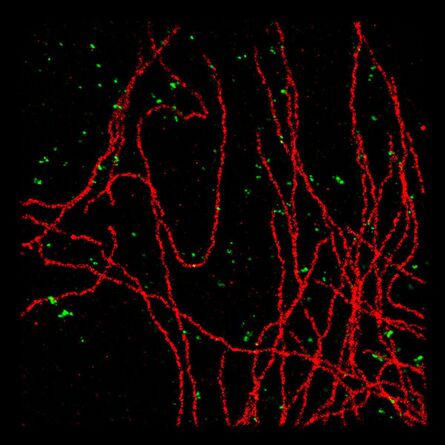
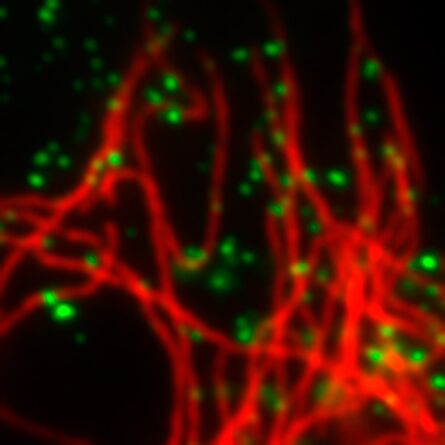
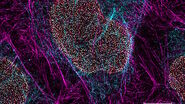
Immunofluorescence
Starting with samples prepared with standard immunofluorescence protocols optimized for diffraction-limited fluorescence microscopy should already provide stunning super-resolution images. To obtain optimal GSD images it might be necessary to increase the concentration of primary and secondary antibodies to achieve a higher labeling density. Increasing the labeling density ensures that the structure of interest is sufficiently stained with fluorophores. A sparse labeling of structures can lead to a point-like appearance of your image. In order to reduce unspecific background signals in the GSD image, it might be necessary to increase the time for blocking your sample as well as the concentration of the blocking reagent. It is advised to wash your samples thoroughly (e.g. by increasing the number and time of washing steps).
For dual color staining Leica Microsystems recommends the combination of a) AlexaFluor 532 with AlexaFluor 647, b) AlexaFluor 555 with AlexaFluor 647 or c) Atto 488 or AlexaFluor 488 with AlexaFluor 647.
Structure preservation
The preservation of a specific cellular structure strongly depends on the fixation method. The exact fixation protocol should be optimized for the structure of interest. For the majority of structures a Paraformaldehyde / Formaldehyde fixation is sufficient. Small amounts of Glutaraldehyde (0.05 % to 0.2 % (v/v)) in addition to Paraformaldehyde / Formaldehyde as a fixant can strongly improve structure preservation. Glutaraldehyde can induce a hazy fluorescent background therefore quenching with ammonium, NaBH4 or other suitable reagents is recommended. Methanol fixation is common for microtubule preservation, but might not be sufficient for other structures.
Coverslips
Super-resolution imaging requires excellent optical performance throughout the whole optical system – not only within the microscope and objective. The coverslip is a major parameter with high impact on imaging results. To obtain optimal super-resolution image quality a flat coverslip is required.
Unfortunately, while being uncritical for standard widefield applications, not all chambered coverslip products can guarantee the degree of flatness optimal for GSD. Furthermore, chambered coverslip products may not provide the necessary mechanical stability for high-resolution imaging. The respective products should be tested by the researcher for their suitability. A better flatness and high stability can be generally achieved by stress free mounting of a glass coverslip, e.g. in specially designed coverslip holders.
In our case, we recommend an alternative method for easy and flat coverslip mounting specifically tailored for GSD imaging utilizing high precision coverslips and a silicone glue to fasten and seal the specimen (see below).
Embedding
The inherent properties of each fluorophore in combination with its direct environment – the embedding medium – determine the ground state depletion capability as well as the single molecule return rate. With it, the embedding medium plays a crucial role for successful GSD imaging and has to be aligned with the utilized fluorophores. Up to now the following growing number of opportunities is applicable.
Embedding medium 1 – MEA in PBS
Reagents
- β-Mercaptoethylamine (MEA) (e.g. Sigma-Aldrich, # 30070, also known as Cysteamine)
- Phosphate Buffered Saline (PBS)
- HCl / NaOH for pH adjustment
Buffer
PBS containing 10 mM β-Mercaptoethylamine (MEA), adjusted to pH 7.4. Dissolve the β-Mercaptoethylamine in PBS and adjust to pH 7.4. The β-Mercaptoethylamine concentration can be varied over a wide range to optimize the GSD image – the usable range is in general between 10 and 100 mM (recommended concentrations for testing 10 / 30 / 100 mM).
β-Mercaptoethylamine solutions must be freshly prepared before use. Alternatively, stock solutions can be prepared and stored at –20°C and freshly thawed before use. The frozen aliquots can be stored for several weeks.
Illumination
EPI: yes
TIRF: yes
| Laser (nm) | Dye | Manufacturer |
|---|---|---|
| 488 | AlexaFluor 488 | life technologies |
| Atto 488 | Sigma | |
| Chromeo 488 | Active Motif | |
| Oregon Green 488 | life technologies | |
| Chromeo 505 | Active Motif | |
| 532 | Atto 520 | Sigma |
| AlexaFluor 532 | life technologies | |
| AlexaFluor 555 | life technologies | |
| AlexaFluor 568 | life technologies | |
| 642 | Atto 633 | Sigma |
| AlexaFluor 647 | life technologies | |
| Atto 647N | Sigma | |
| Atto 655 | Sigma | |
| AlexaFluor 680 | life technologies | |
| AlexaFluor 700 | life technologies |
Embedding medium 2 – Glucose Oxidase
Reagents
- Glucose Oxidase (e.g. Sigma-Aldrich G2133)
- Catalase (e.g. Sigma-Aldrich C1345)
- Glucose
- Phosphate Buffered Saline (PBS)
- HCl / NaOH for pH adjustment
Buffer
PBS containing 10 % (w/v) glucose, 0.5 mg/mL glucoseoxidase and 40 μg/mL catalase adjusted to pH 7.4.
Note: It is recommended to prepare a glucose stock solution and mix the reagents freshly before mounting the coverslip.
Illumination
EPI: yes
TIRF: yes
| Laser (nm) | Dye | Manufacturer |
|---|---|---|
| 532 | Atto 532 | Sigma |
| AlexaFluor 532 | life technologies | |
| AlexaFluor 555 | life technologies | |
| Rhodamine 6G | Active Motif | |
| Atto 565 | Sigma | |
| AlexaFluor 568 | life technologies | |
| 642 | Atto 655 | Sigma |
| AlexaFluor 680 | life technologies |
Embedding medium 3 – PVA
Reagents
- Polyvinyl alcohol (PVA) with a molecular weight of 25,000, 88 mol% hydrolyzed (e.g. Polysciences, #02975-500)
- Phosphate Buffered Saline (PBS)
- HCl / NaOH for pH adjustment
Hardware
Spincoater
Buffer
Prepare a solution of 1 % PVA in PBS. Adjust the pH to 7.4.
Spincoating
Place the coverslip on the spincoater and dry the coverslip with brief 5–10 sec spin at approx. 3,000 rpm. Drop 50 μl of the PVA-solution onto the sample and spin the sample at approx. 3,000 rpm for approximately 30 s. Let the PVA-film dry for a few minutes and mount the sample. The sample can be stored at room temperature and used for GSD imaging for up to three months.
No additional mounting medium is required. The PVA-film is protecting the sample.
Illumination
EPI: yes
TIRF: not possible
| Laser (nm) | Dye | Manufacturer |
|---|---|---|
| 488 | AlexaFluor 488 | life technologies |
| Atto 488 | Sigma | |
| Chromeo 488 | Active Motif | |
| Oregon Green 488 | life technologies | |
| Atto 520 | Sigma | |
| 532 | AlexaFluor 532 | life technologies |
| AlexaFluor 680 | life technologies |
Embedding medium 4 – Mowiol
Reagents
- Glycerol (analytical grade)
- Mowiol 4-88 (e.g. Calbiochem #475904)
- Distilled water
- TRIS
- HCl / NaOH for pH adjustment
Embedding medium
- Start with 6 g Glycerol
- Add 2.4 g Mowiol 4-88
- Add 6 ml Aqua dest.
- Add 12 ml 0.2 M TRIS buffer pH 8
- Stir for 4 hours (magnetic stirrer)
- Let suspension rest for 2 hours
- Incubate for 10 min at 50 °C (water bath)
- Centrifuge at 5,000 x g for 15 min
- Take the supernatant and freeze it in aliquots at –20 °C
Illumination
EPI: yes
TIRF: not possible
| Laser (nm) | Dye | Manufacturer |
|---|---|---|
| 488 | Atto 488 | Sigma |
| Chromeo 505 | Active Motif | |
| 532 | AlexaFluor 532 | life technologies |
| Rhodamine 6G | Active Motif | |
| Atto 647N | Sigma | |
| 642 | Atto 655 | Sigma |
| AlexaFluor 700 | life technologies |
Embedding medium 5 – Vectashield®
Reagents
- Vectashield® (e.g. Vector Laboratories, # H-1000)
- 50 mM Tris in Glycerol
- HCl / NaOH for pH adjustment
Buffer
- 50 mM Tris/Glycerol buffer containing 10 % Vectashield® adjusted to pH 7.4.
- Dissolve Tris in Glycerol to prepare a 50 mM stock solution. Mix 10 % Vectashield® with 90 % Tris/Glycerol buffer. Adjust to pH 7.4. Samples can be stored over several weeks at 4 °C.
Illumination
EPI: yes
TIRF: not possible
| Laser (nm) | Dye | Manufacturer |
|---|---|---|
| 532 | AlexaFluor 555 | life technologies |
| 642 | AlexaFluor 647 | life technologies |
Embedding medium for YFP
GSDIM is suitable to create super-resolution images using fluorescent proteins. E.g. the fluorescent protein YFP / Venus displayed astonishing results. Furthermore, other fluorescent proteins should work with this technology (including GFP). To combine the power of organic fluorophores with the advantages of genetically encoded fusion proteins (e.g. if no suitable antibody is available), the use of tags like SNAP-, Halo- or CLIP-Tag is recommended. These tags can be genetically linked to the protein of interest and later on labeled with organic fluorophores (e.g. after fixation and permeabilization).
The following aqueous embedding was successfully used to image YFP in fixed and permeabilized mammalian cells:
- Catalase SIGMA, C1345 40 ug/mL
- Glucose Oxidase SIGMA G2133 0.5 mg/mL
- Glucose 10 mg/mL
- β-Mercaptoethylamine (Cysteamine) 1 mM
- Diluted in PBS, adjusted to pH 7.4
Mounting
Leica Microsystems recommends mounting the coverslip with the sample directly on depression slides. The coverslip should be fixed to the depression slide and sealed with the two-component silicone-glue Twinsil® (Picodent, Wipperfürth, Germany, #13001000). Twinsil® is non-toxic, fast hardening and can be later on easily removed, e.g. to re-mount the sample.
- Add 90 μl of aqueous embedding medium (MEA, Glucose-Oxidase-Mix or Vectashield®) into the cavity of a clean depression slide.
(For embedding media PVA and LR-White: leave the cavity of the depression slide empty.) - Place the glass coverslip with your sample onto the cavity of the depression slide. The sample should face the cavity. The coverslip should cover the depression completely.
- If aqueous embedding medium is filled in the cavity, ensure that no air bubbles are present. Otherwise carefully remove the coverslip and repeat the procedure.
- No liquid should be present at regions, which should be covered by the glue (Twinsil®). Carefully remove remaining liquid with filter paper. For Embedding Media 1, 2 and 5: Ensure that no buffer is soaked out from the reservoir.
- Mix the yellow and blue component of Twinsil® in a quantity of 1+1 thoroughly and apply it to the edges of the glass coverslip without directly touching the coverslip.
For aqueous embedding media: Seal the coverslip completely with Twinsil®.
For embedding media PVA and LR-White: Seal the coverslip to approx. 75 % and leave the remaining quarter open to allow ventilation and avoid condensation on the sample. - After 5–10 minutes the two-component glue is hardened and the sample is ready for GSD imaging.
The glue can be easily removed without leaving traces on the glass after hardening (e.g. to exchange the imaging buffer).
AlexaFluor® is a registered trademark of life technologies™.
Mounting procedure
1) Pipet approx. 90 µl of liquid embedding medium (e.g. MEA solution) into the cavity of the depression slide. |  | ||
2) Place the round coverslip – holding the immunostained sample – onto the medium and avoid the formation of air bubbles. |  | ||
3) Gently press onto the coverslip to remove air bubbles (if necessary) and excess of buffer. |  | Mix the two components – hardening of Twinsil® starts with mixing. | |
4) Carefully remove liquid without soaking buffer from the reservoir in the depression slide. |  |  |  |
5) Seal the coverslip by carefully applying Twinsil®. Do not touch or move the coverslip! |  |  | |
6) When the hardened (approx. 5 min) Twinsil® is touched gently with a tip, it will not stick nor can it be pushed aside. |  |
Imaging tips
Dual-color stainings
Leica Microsystems recommends the combination of
a) AlexaFluor 532 with AlexaFluor 647
b) AlexaFluor 555 with AlexaFluor 647
c) Atto 488 or AlexaFluor 488 with AlexaFluor 647
for sequential dual color imaging. Please image the red channel (AlexaFluor 647) first. Leica Microsystems does NOT recommend imaging the green/orange channel (e.g. AlexaFluor 532) at the beginning to avoid bleaching of AlexaFluor 647, which can be – although at very low levels – excited by a 488 nm (or 532 nm) laser. If it is intended to use other fluorophores for double-staining of the sample, please check that both fluorophores can be imaged under the same GSD imaging conditions (embedding media) and crosstalk is negligible.
Use of 405 nm laser
The Leica SR GSD 3D is equipped with a 405 nm laser. The lifetime of fluorophores in the off state can be shortened by illuminating the sample additionally with UV-light. The shorter lifetime leads to an increase of fluorophores in the on state and therefore can be used to increase the number of detected events per frame. By keeping the number of events per frame in an optimal range, it is possible to a) efficiently detect well-separated single fluorophores and b) keep the time to acquire a super-resolution image short, because it is possible to accumulate the required number of events within fewer images.
When acquiring the longer wavelength channel of dual-color images, it is important to use the 405 nm laser carefully! The 405 nm wavelength can excite green dyes and therefore bleach the shorter wavelength fluorophores while acquiring the longer wavelength super-resolution image. The optimal settings should be determined for each sample type separately.
Super-resolution imaging of living cells
Recent reports have studied the suitability of GSDIM (also known as dSTORM) to image living cells. In general, it is possible to study living cells with the Leica SR GSD 3D, but phototoxicity – as a result of the high laser power applied – and the dynamics of structure of interest during the observation period should be carefully controlled.