Opto-digital image processing tools, such as Computational Clearing [2], can help to improve the signal-to-background ratio and reduce the out-of-focus blur. The associated contrast enhancement can reveal additional information in microscope images.
Immunofluorescence of viral proteins
Amongst sequencing techniques, bioinformatics, and electron microscopy, Ogando et al. [1] analyzed infected cells by fluorescence microscopy: Vero E6 cells were grown on glass cover slips, infected with SARS-CoV-2, and fixed with paraformaldehyde. Then, the cells were incubated with antisera from rabbits or mice which were exposed to SARS-CoV beforehand (Figure 2). The SARS-CoV-originated antibodies, which bind to SARS-CoV-2 structures in Vero E6 cells, were then detected by fluorescently labelled secondary antibodies. In addition, nuclei were stained with Hoechst. Fluorescence imaging was done with a DM6 B upright fluorescence microscope.
SARS-CoV antisera cross-react with SARS-CoV-2
Immunofluorescence microscopy revealed cross reactivity of many SARS-CoV antisera in SARS-CoV-2 infected cells (viral proteins nsp3, nsp4, nsp5, nsp8, nsp9, nsp13, nsp15, N, M). This fact means that antisera produced against SARS-CoV also lead to characteristic fluorescent staining in SARS-CoV-2-infected cells (Figure 3). Whereas nsps proteins were found in the perinuclear region of infected cells, the N protein was spread throughout the cytosol. The M protein was detected in the Golgi apparatus.
Potential of upright fluorescence microscopy for virology research
The cross-reacting antisera described in the study of Ogando et al. [1] is a useful tool for the characterization of the replication cycle of SARS-CoV-2. This tool enables researchers to define potential targets for inhibitors of replication.
A relatively simple experimental setup – immunofluorescence microscopy – is sufficient to draw conclusions of the viral replication cycle. Because the cells are grown on cover slips for this method and mounted on glass slides, an upright fluorescence microscope is a practical solution. Automated versions with a motorized stage in combination with a large field of view (FOV) help users to acquire large overviews quickly. If single snapshot images are enough, a manually operated mechanical stage is the more reasonable choice.
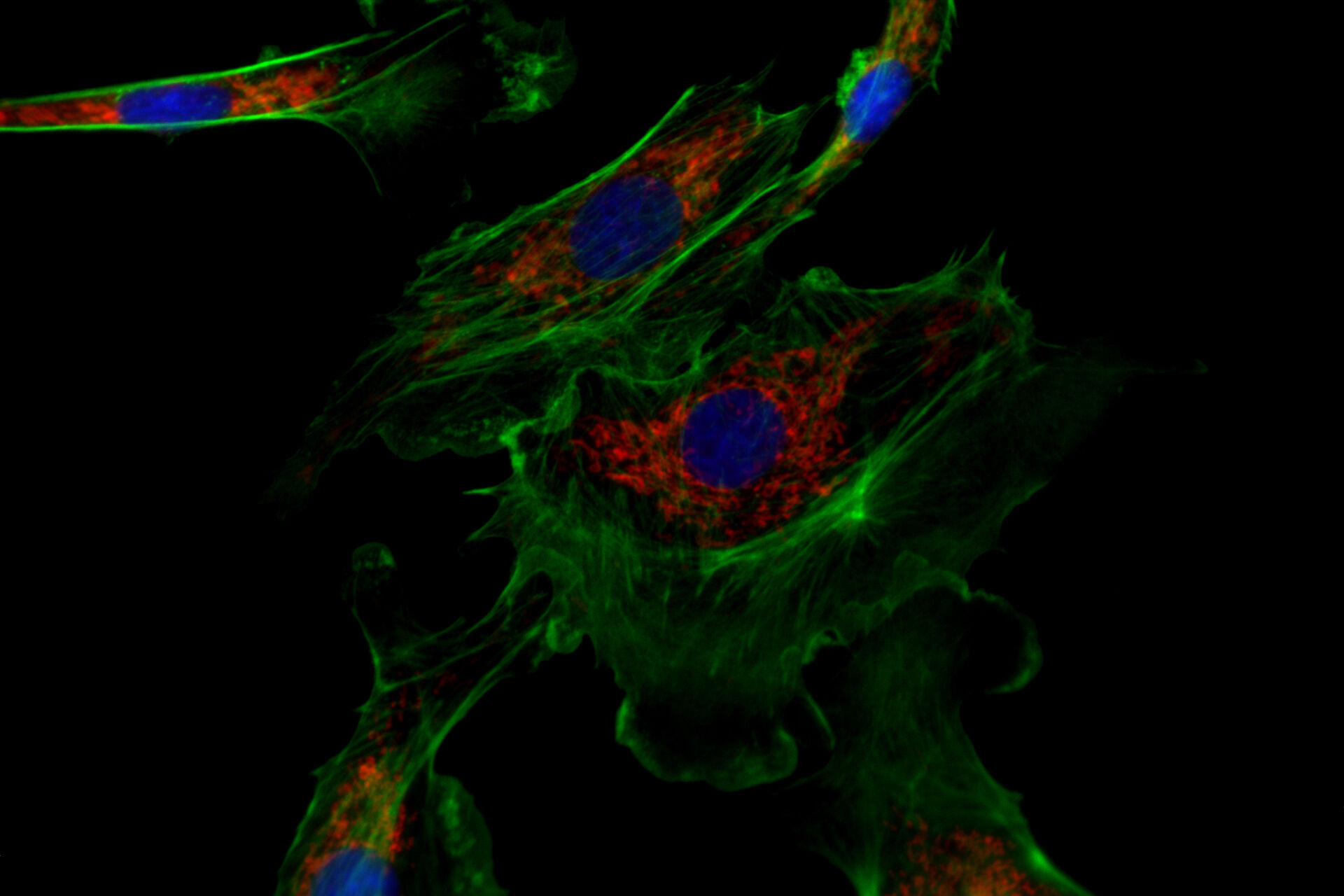
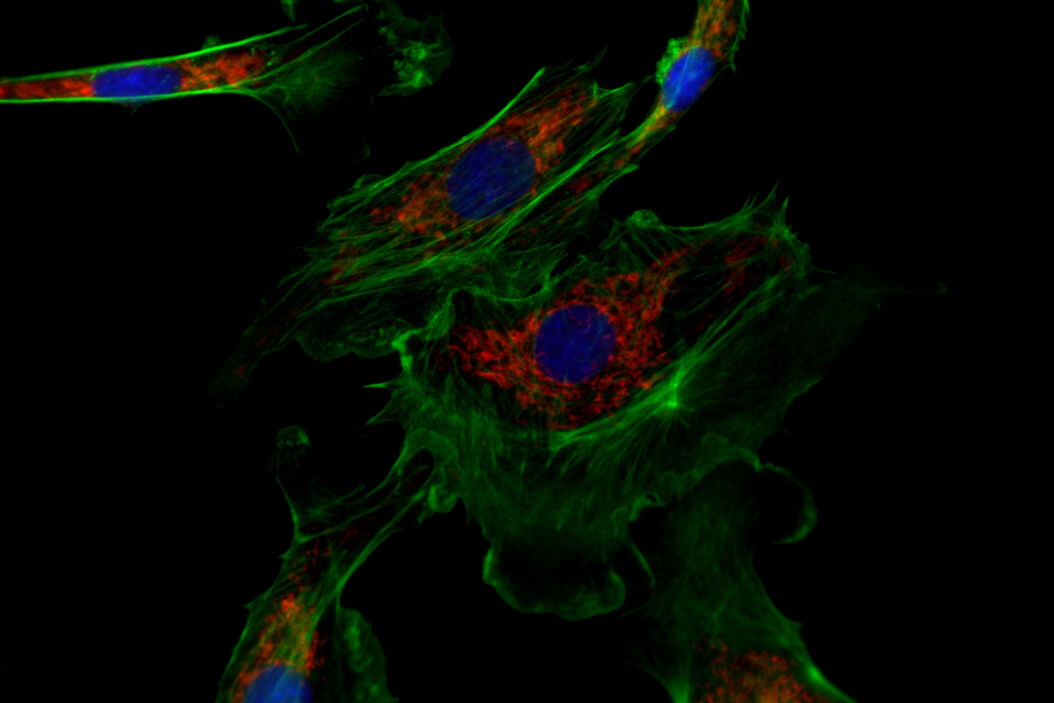
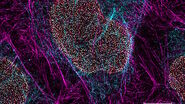
![One of the experiments of Ogando et al. [1] involved the use of immunofluorescence microscopy to image Vero E6 cells. One of the experiments of Ogando et al. [1] involved the use of immunofluorescence microscopy to image Vero E6 cells. In principle, they explored the potential of antisera which have been produced against SARS-CoV in rabbits or mice during the outbreak in 2002 and 2003. They used different rabbit antisera as a 1st set of antibodies. Then, these rabbit antisera were labeled with a 2nd set of antibodies which were fluorescent. Subsequently, fluorescence microscopy was performed with a DM6 B microscope.](/fileadmin/_processed_/f/c/csm_Vero_E6_cells_imaged_with_immunofluorescence_microscopy_Ogando_Serum_1bf2c697b4.png)
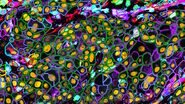
![Immunofluorescence microscopy of SARS-CoV-2 infected Vero E6 cells which were treated with rabbit SARS-CoV antisera. Immunofluorescence microscopy of SARS-CoV-2 infected Vero E6 cells which were treated with rabbit SARS-CoV antisera [1]. Nsps proteins can be found near the nucleus, while the M protein is located at the Golgi apparatus. Anti-dsRNA antibodies were used to label replicative intermediates of viral RNA synthesis. Nuclei were stained with Hoechst. Scale bar is 25 μm for 3a and 3b and 100 μm for 3c, 3d, and 3e.](/fileadmin/_processed_/a/4/csm_Cross-reactivity_of_antisera_SARS-CoV_proteins_c7b9ba4f6c.jpg)