Bacterial infection in the mouse mucosal barrier
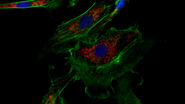
Jarret et al. did not investigate viral but rather bacterial infection in the mouse mucosal barrier. By using single-molecule fluorescence in situ mRNA hybridization (smFISH; THUNDER Imager 3D Live Cell) they discovered that intestinal neurons produce the cytokine IL-18. With it, they could show that neuron derived IL-18 signaling plays a major role in intestinal immunity which was not yet discovered.
Monitor markers for immunosuppressive mechanisms
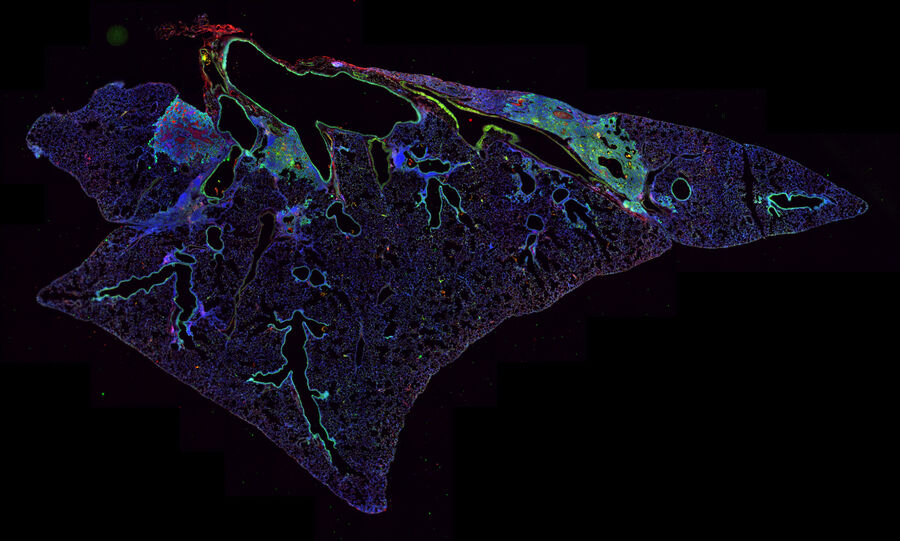
Immunosuppression can be studied with the help of viral infected mouse models. Andrew Beppu used large multi-channel tile scans of mouse lungs to monitor markers for immunosuppressive mechanisms. The mice were infected with the influenza virus and then investigated for regression of lung basal-like structures (THUNDER Imager 3D Cell Culture).
DENV induced gene expression effects
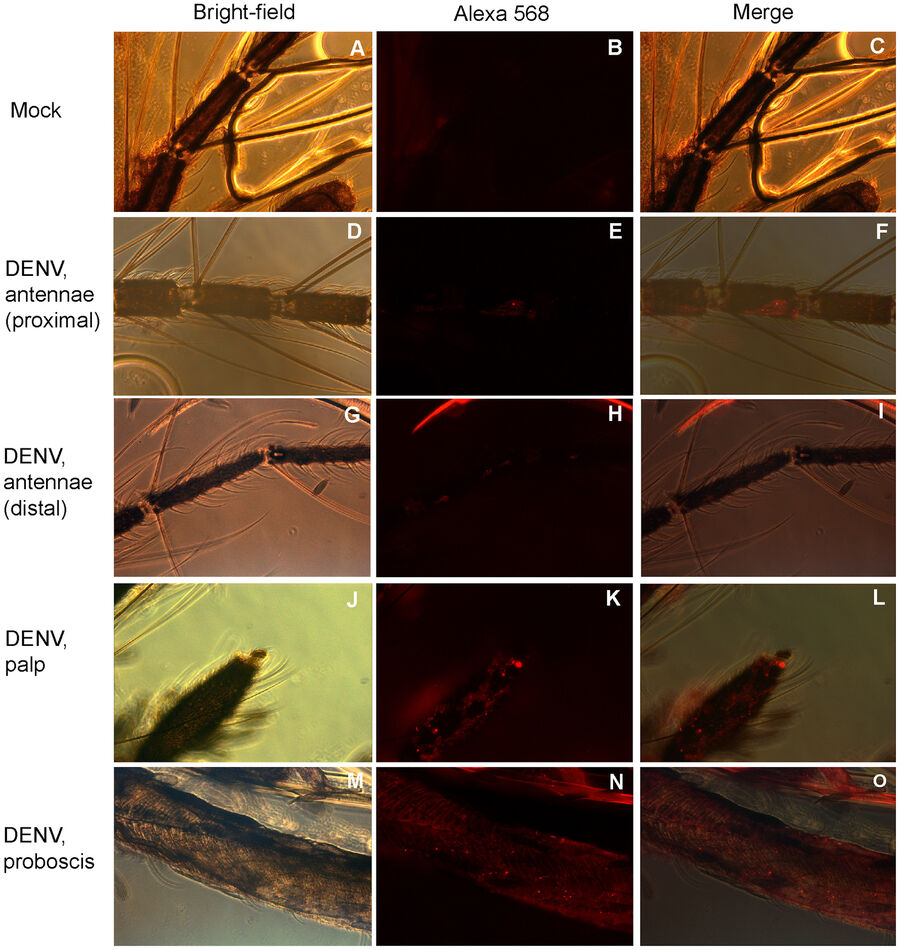
Sim et al. studied Dengue virus (DENV) induced gene expression effects in the salivary gland of the yellow fever mosquito (Aedes aegypti). The authors found out that upon DENV infection, the transcriptome of the mosquito salivary gland was changed. By using microarrays for gene expression analysis, they could show for the first time that some genes were expressed which have influence on the mosquito’s host-seeking and probing behavior. They used immunofluorescence microscopy to identify regions in the mosquito’s chemosensory organs where the virus was abundant (see Fig. 3).
https://journals.plos.org/plospathogens/article?id=10.1371/journal.ppat.1002631#s4
The morphology of Hendra virus infected cells
Monaghan et al. characterized the morphology of Hendra virus infected cells with the help of microscopy. Confocal images (SP5) revealed the intracellular viral protein distribution, whereas super-resolution microscopy (SR-GSD) even gave a closer look into the protein distribution inside virions.
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4254186/
Cellular RNA helicase binding to La Crosse virus (LACV) nucleocapsids
Super-resolution microscopy was utilized by Weber et al. to study cellular RNA helicase binding to La Crosse virus (LACV) nucleocapsids (SR-GSD). For this purpose, immunofluorescence was applied to stain the viral LACV N protein and cellular RNA helicase RIG-I in infected A549 cells. Super-resolution microscopy supported biochemistry data that RIG-I binds the nucleocapsids via the 5’ppp dsRNA panhandle.
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5515363/
Natural killer (NK) cells
Maze and Orange investigated natural killer (NK) cells which are important counterplayers of viruses. They belong to the innate immune system and surveil virally infected and tumorigenic cells. Directed secretion of specialized secretory lysosomes via the immunological synapse kills the virally infected cells. STED nanoscopy (TCS SP8 STED) was used to decipher the interaction of lytic granules with the cytoskeleton of the NK cells.
Image the immunological synapse with STED nanoscopy
Another publication by Zheng et al. describes the technical procedure which can be used to image the immunological synapse with STED nanoscopy (TCS SP8 STED).
https://www.jove.com/video/52502/super-resolution-imaging-natural-killer-cell-immunological-synapse-on
Shed light on parasitology problems with STED nanoscopy
In addition to virology and immunology, STED nanoscopy can also shed light on parasitology problems, such as merozoite invasion in erythrocytes. 3D STED (TCS SP8 STED) reveals spatial information of the protein components which are involved in Plasmodium infection.
Influence of coronaviruses on cellular NF-κB signaling and chromatin landscape
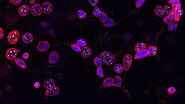
Poppe et al. investigated the influence of coronaviruses on cellular NF-κB signaling and chromatin landscape. With the help of Laser Microdissection (LMD6000), they isolated cells expressing the coronavirus N protein and extracted their entire RNA. By utilizing RT-qPCR and microarray analysis, they found out which mRNAs were over- or under-represented in the cells. Furthermore, they used immunofluorescence of the coronavirus N protein to monitor viral infection and its spread in A546 cells (DMIRE2, DMi8).
https://journals.plos.org/plospathogens/article?id=10.1371/journal.ppat.1006286#sec002)
Brightfield microscopy plays a direct, but only minor role in virology, because fluorescence microscopy fulfills better the demands of researchers. Nevertheless, there are opportunities for brightfield microscopy when animal tissues are investigated. For example, researchers can check tissues for morphology changes after viral infection, etc.
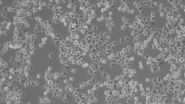
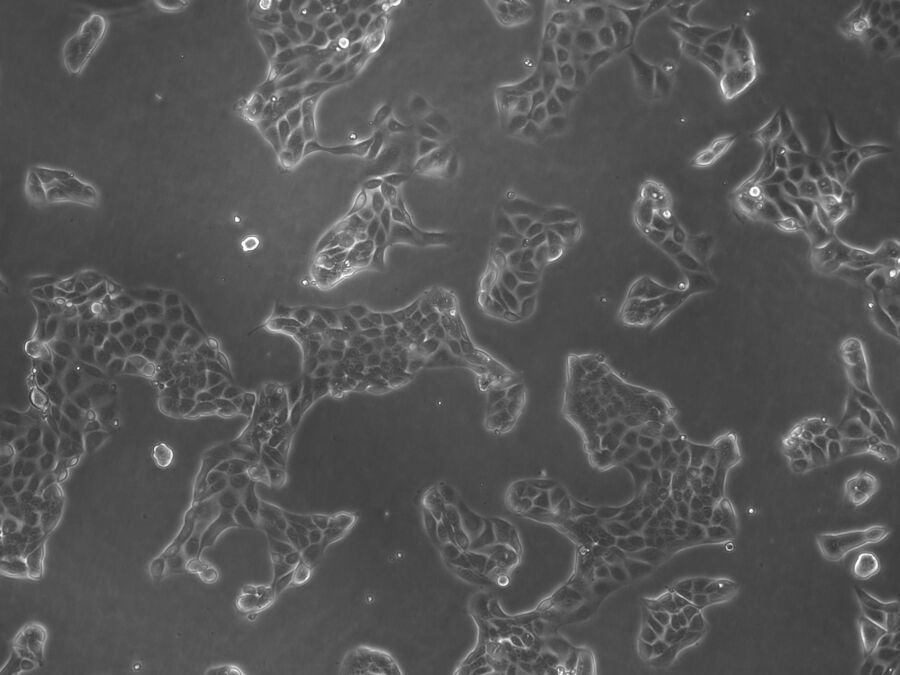
In addition, brightfield microscopy is used in cell culture labs to check the health and growth status of cells (see Fig. 5) which are already infected or will be at a later time (DM IL, DMi1).
This list of microscopy methods is not meant to be comprehensive. There are other techniques which are also used to visualize viruses, but are not in the scope of this article. For example, electron microscopy (EM) can resolve virus particles. Single molecule detection and Fluorescence Lifetime Imaging (FLIM) (STELLARIS 8 FALCON), as well as multiphoton microscopy (SP8 DIVE) are additional methods, which are suitable for virology (see references).
Fig. 6: Sample preparation for electron microscopy. Image taken from https://www.youtube.com/watch?v=dtKT0FUKKxA at EMBL, Heidelberg.
SARS-CoV and SARS-CoV-2 were able to infect enterocytes
Lamers et al. investigated, if the coronavirus SARS-CoV-2 infects not only the respiratory system, but also the human gut. By using confocal microscopy (SP8) and electron microscopy (EM sample preparation: EM UC7) with human small intestine organoids, they could show that SARS-CoV and SARS-CoV-2 were able to infect enterocytes.
To find out more about electron microscopy, please refer to the examples given in the references below.
References
Confocal Microscopy in Virology
Visualizing Oncolytic Virus-Host Interactions in Live Mice Using Intravital Microscopy
Inhibition of Cytosolic Phospholipase A2α Impairs an Early Step of Coronavirus Replication in Cell Culture
Characterization of Virus-Specific Vesicles Assembled by West Nile Virus Non-Structural Proteins
Host Sphingomyelin Increases West Nile Virus Infection in Vivo
Fluorescence Lifetime Imaging (FLIM) in Virology
Quantitative FRET-FLIM-BlaM to Assess the Extent of HIV-1 Fusion in Live Cells
Single Molecule Detection (SMD) in Virology
A Dynamic Three-Step Mechanism Drives the HIV-1 Pre-Fusion Reaction
Toremifene Interacts With and Destabilizes the Ebola Virus Glycoprotein
Dynamin-2 Stabilizes the HIV-1 Fusion Pore With a Low Oligomeric State
Astrocytes Resist HIV-1 Fusion but Engulf Infected Macrophage Material
Multiphoton Microscopy in Virology
Single-cell glycolytic activity regulates membrane tension and HIV-1 fusion
Widefield Microscopy in Virology
Super-Resolution Microscopy in Virology
Laser Microdissection in Virology
Electron Microscopy in Virology
SARS-CoV-2 productively infects human gut enterocytes
EMBL scientists using electron microscopy to look at coronavirus infected cells
Other Infectious Agents
Observing Malaria Infection at the Right Spot in the Human Host
Inflammation
Chronic Inflammation Under the Microscope
Cell Culture
Introduction to Mammalian Cell Culture - Morphology and Cell Types & Organization