Development of the central nervous system (CNS)
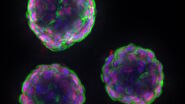
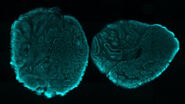
One area of research in developmental biology involves the study of the assembly of neural circuits, their activity, and the formation of the central nervous system (CNS) during the development of an organism [1]. One model organism used for this type of investigation is the Drosophila-melanogaster fruit fly [2]. One way to visualize neural activity in Drosophila embryo is via expression of the GCaMP, a potent genetically encoded calcium indicator (GECI), in the fly embryo’s neurons [1]. Short bursts of GCaMP emission can be seen with time-lapse imaging sequences and help provide a better understanding of the effect of calcium transients on the model organism’s life [1].
Challenges imaging neuronal activity in embryos
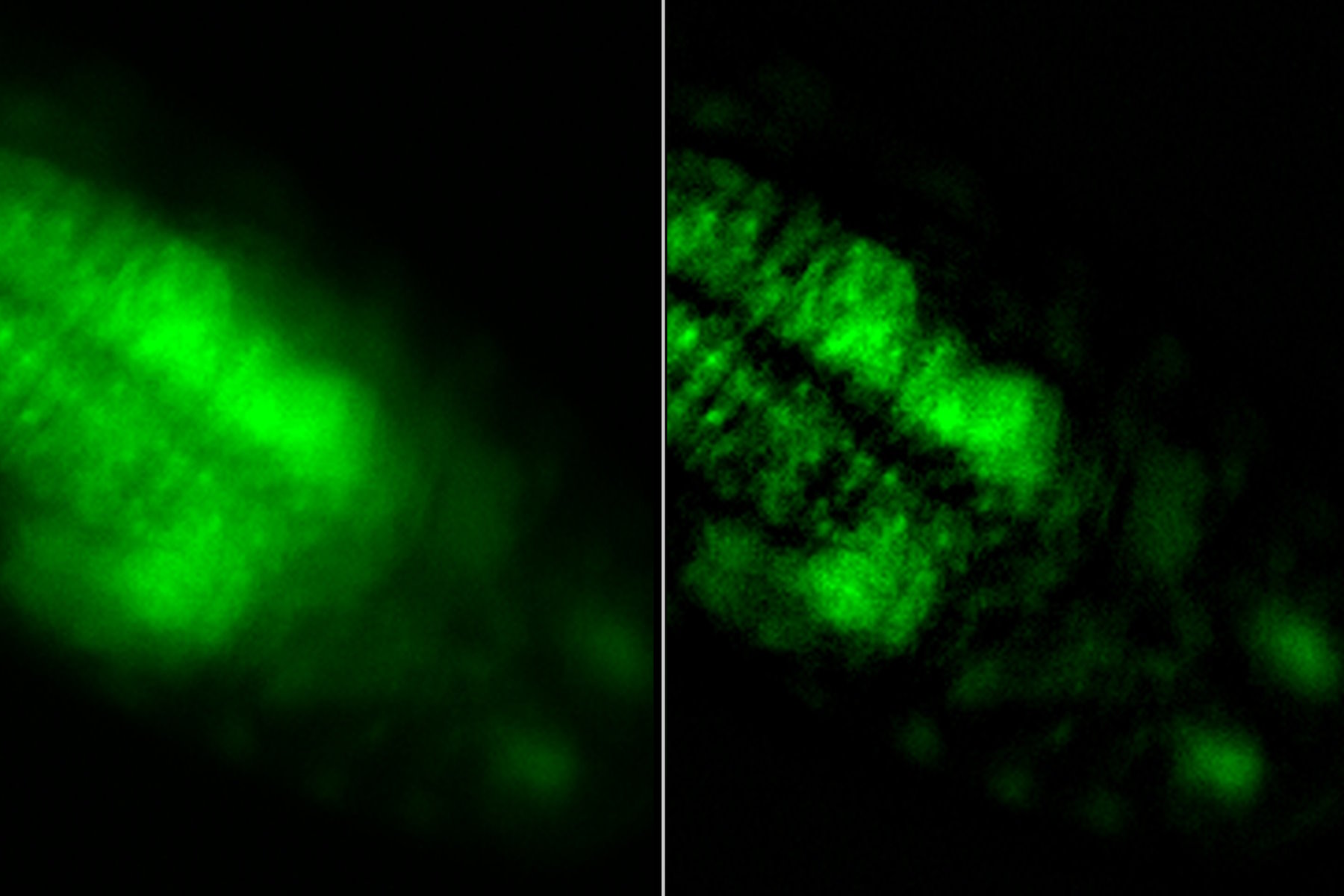
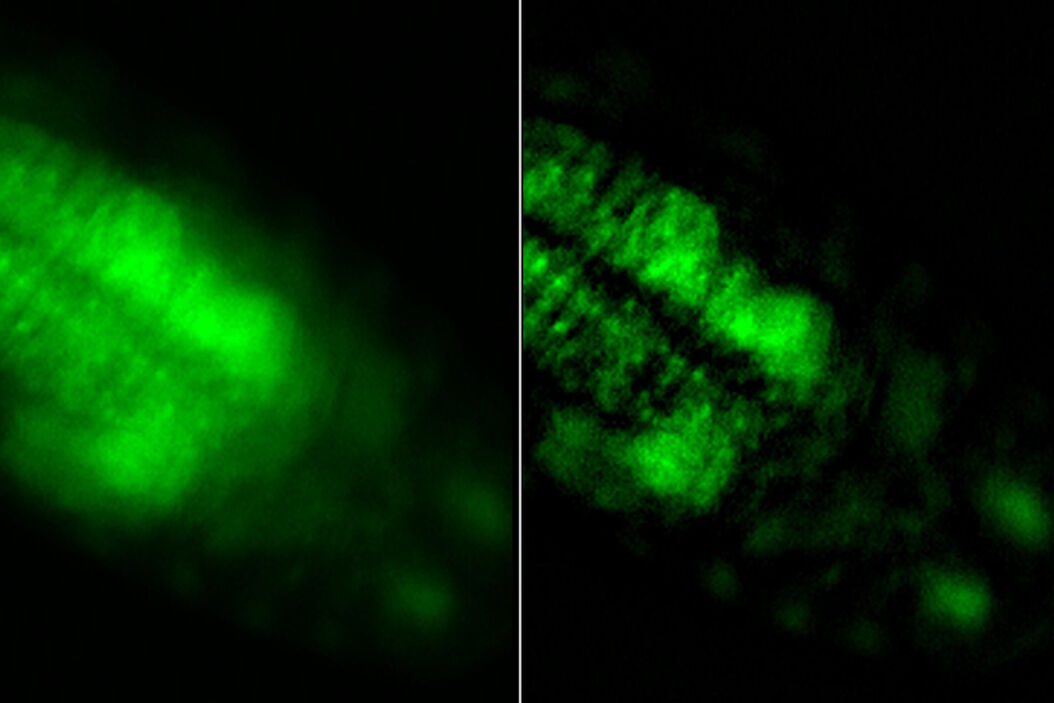
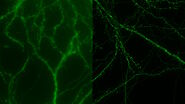
A significant challenge that often arises when imaging thick specimens with widefield fluorescence microscopy, such as whole embryos of Drosophila fruit flies, is the out-of-focus blur or haze produced by light scattering [4,5]. Such haze can it make difficult to resolve structures of interest inside the embryo.
Methods to investigate whole embryos
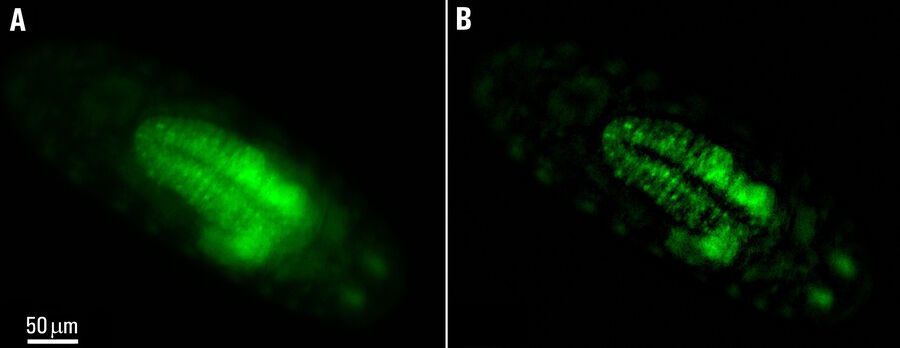
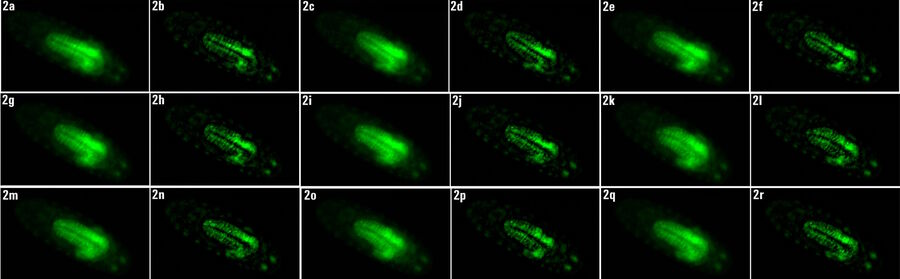
Whole embryos of Drosophila-melanogaster fruit flies expressing the GCaMP calcium indicator in their neurons were used as specimens. Imaging of the embryos, while maintained under proper physiological conditions, was performed with a THUNDER Imager Model Organism using a 1x objective lens. Instant Computational Clearing (ICC) was applied to remove out-of-focus blur or haze common for raw widefield images. Time-lapse imaging was done to investigate calcium transients in the embryo neurons via GCaMP-emission bursts. The original data set was acquired for approximately 4.5 minutes and each image taken with an exposure time of < 400 ms.
Results
Images of a Drosophila embryo expressing GCaMP in its neurons are shown in figure 1. A time-lapse image series illustrating calcium transients, via GCaMP bursts in neurons of a Drosophila embryo, is shown in figure 2. The average time interval between displayed images is 10.47 seconds. The left column shows raw widefield images and the right column THUNDER/ICC processed images.
Conclusions
The results presented here show how the investigation of central nervous system (CNS) development and neural activity in whole Drosophila-melanogaster embryos can be improved with a THUNDER Imager in comparison to conventional widefield microscopy. The embryos expressed a GCaMP calcium indicator in their neurons. A time-lapse image series showing GCaMP bursts due to calcium transients in the embryo neurons was obtained. These results may help provide a better understanding of the effects of calcium transients on the embryo’s neural development.