That is why Leica LMD Systems allow for collection directly into your downstream analysis containers, such as PCT microtubes for fast tissue homogenization, protein extraction, and digestion .
Pressure Cycling Technology (PCT, PressureBioSciences Inc.) uses high pressure to streamline sample preparation for mass spec analysis. PCT can process LMD samples in one hour, and larger biopsy tissue pieces in under three.
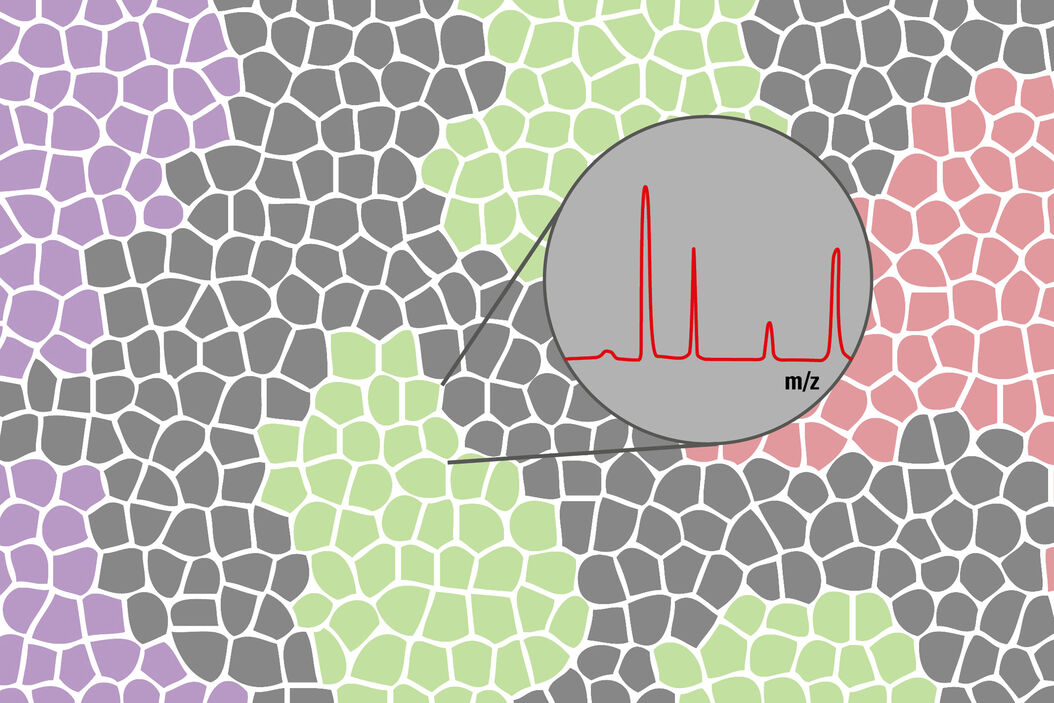
The combined techniques allow for separation of the tumor microenvironment with high precision and speed. Downstream molecular analysis of the different regions will be meaningful, since they can be analyzed separately and not as a mixture.
Download Flyer
Biomarker Discovery Workflow with Single Cell Precision
Leica's Laser Microdissection systems will improve your workflow by precisely cutting only the cells you are interested in. Everything under visual control and without contamination with surrounding tissue.
Download: Flyer Biomarker Discovery
1. Sample preparation

Typically, special membrane-based slides are used for laser microdissection. Sample preparation for LMD is straightforward, and can be derived from classical preparations for histology.
For downstream proteomics we recommend PET membrane slides (which are almost free of softeners) or DIRECTOR™ slides which allow completely membrane-free ablation.
Paraffin embedding brings the tissue sample into a constitution which can be cut into thin slices. Alternatively you can utilize cryo-sectioning to prepare your slides.
We will deliver the suitable LMD slides for your application.
2. Fixation and staining

Fixation and staining of tissue for LMD application can be applied according to the structures of interest. This can comprise both, brightfield and fluorescence stains.
This step can be automated with a stainer.
In general, most stains are compatible with downstream proteomics. Nevertheless, it is worth investigating before.
3. Visualization and ROI

Leica LMD systems can generate an automatic sample overview that can be used to easily navigate to your regions of interest (ROI).
Your ROIs can be identified and marked automatically by the ADM software module, or you can apply shapes manually.
- More information on the Leica LMD systems
- Laser microdissection articles & tutorials
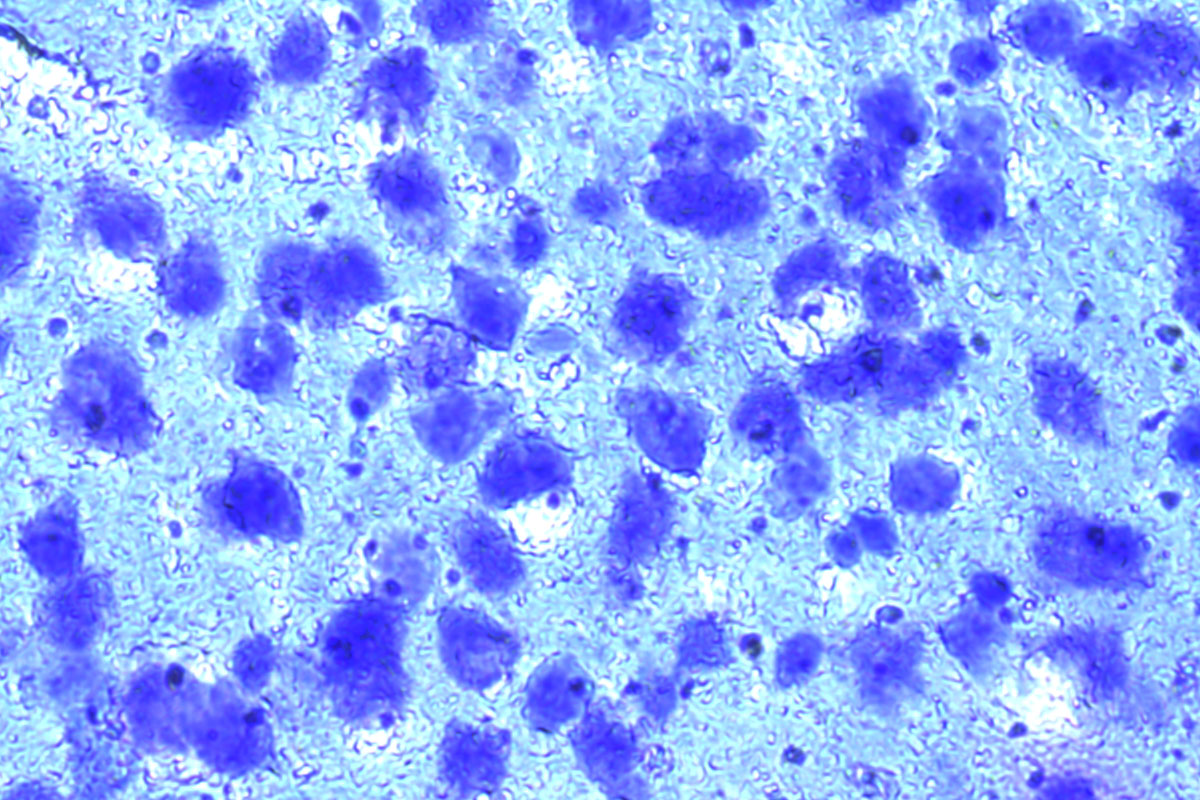
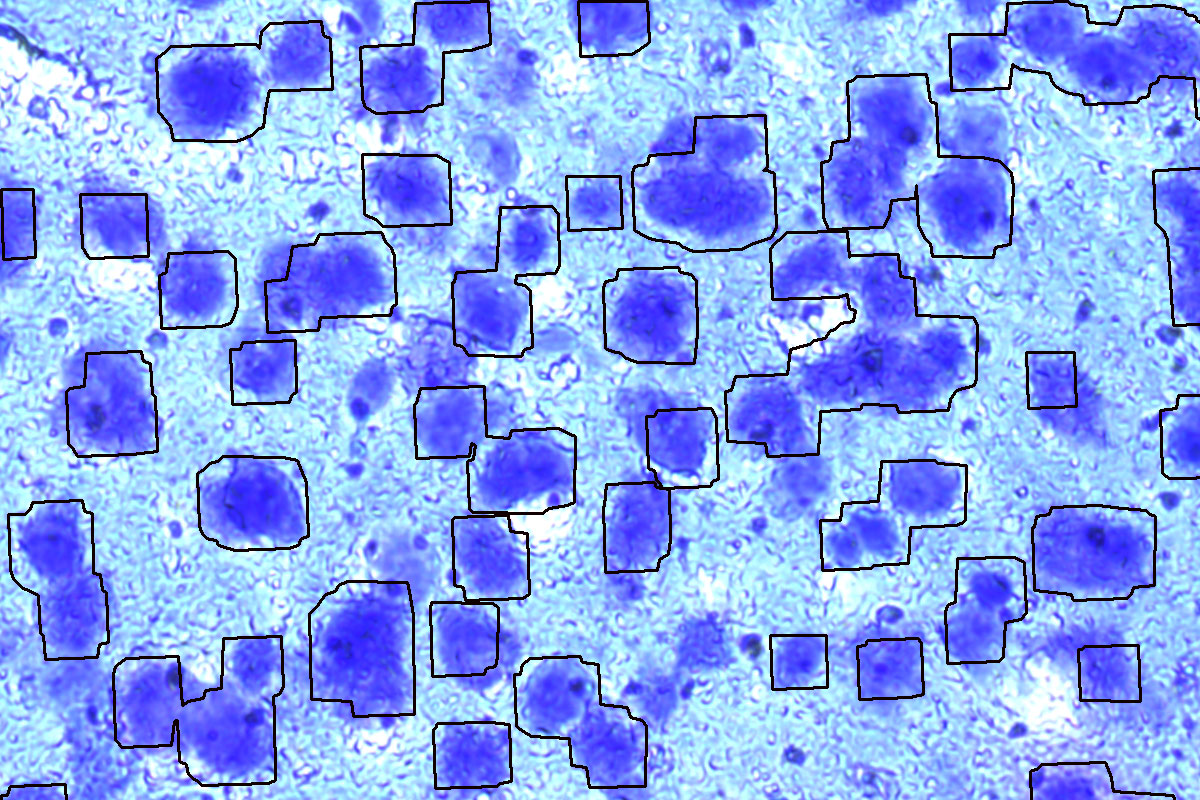
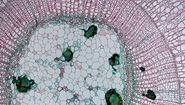
Mouse brain, Cresyl violet. Before and after auto detection (ADM) of neurons.
4. Laser microdissection

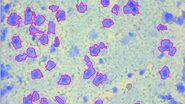
Next, the defined ROIs are dissected by the laser under visual control and directly collected via gravity.
Isolate cancer material or any other ROI with a few mouse clicks and pool – if necessary – in one or different reaction vessels if desired.
You can use standard consumables, such as PCR tubes. Or collect them directly into your downstream analysis vessel, e.g. PCT μTubes (Pressure BioSciences Inc.) with a dedicated LMD collection device, see next step.
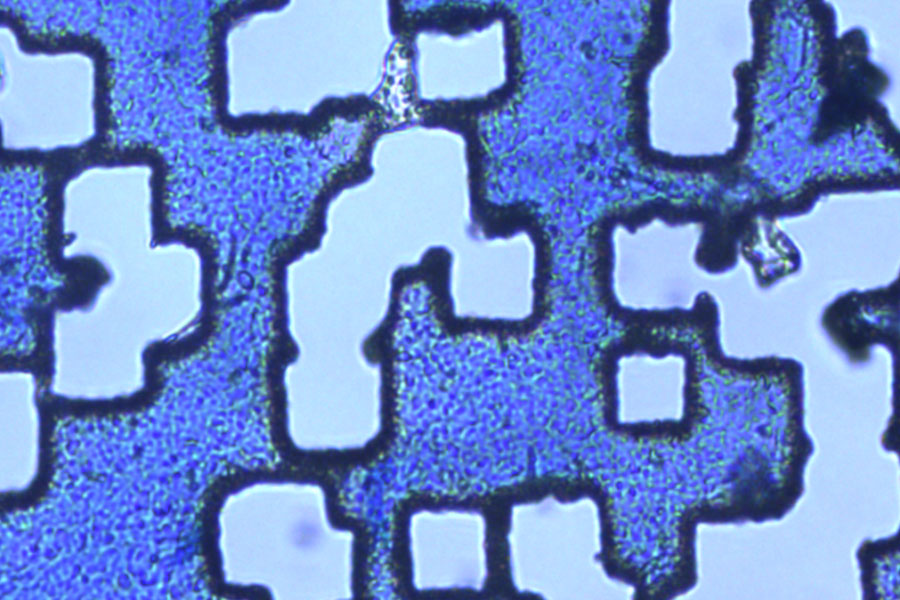
5. Protein preparation
Homogenization, extraction and digestion of minute samples can be done e. g. with Pressure Cycling Technology (PCT) from Pressure BioSciences Inc. Following the LMD-PCT workflow this allows for rapid and reproducible preparation of peptides in just over one hour (Lee S et al., Cell Reports 2020), saving several hours, and providing hands off sample preparation for minute tissue samples.

6. Analysis
Analyze your samples by mass spectrometry methods and identify your biomarkers.
Typical fields of research
- Biomarker Discovery
- Cancer Research
- Personalized Medicine
- Translational Research
- Proteomics
- Metabolomics
References
- Lee S et al., Cell Reports 2020, https://doi.org/10.1016/j.celrep.2020.03.066
- Hunt AL et al., Cancer Res 2019;79 doi.org/10.1158/1538-7445.AM2019-4709
- Guo T et al., Nat Med. 2015; 21:407-13
- Shao S et al., Proteomics 2015; 15: 1-11