RNA Analysis Workflow with Single Cell Precision
Laser microdissection (LMD) systems from Leica Microsystems can improve your workflow by enabling you to precisely cut only the cells you are interested in. Everything remains under visual control and without contamination by surrounding tissue.
RNA Analysis Workflow - Steps
1. Sample preparation

Typically, special membrane-based slides are used for laser microdissection. Sample preparation for LMD is straightforward and can be derived from classical preparations for histology. Paraffin embedding gives the tissue sample a better consistency for cutting it into thin slices. Alternatively, you can utilize cryo-sectioning to prepare your slides. Tissue for RNA analysis is often flash-frozen, e.g., in liquid nitrogen, and then cryo-sectioned.
Leica Microsystems can provide you with LMD slides which are suitable for your application .
2. Fixation and staining

The cresyl violet stain is commonly used for neuronal tissue. It is a basic stain that binds to acidic molecules of neuronal cytoplasm, such as, RNA-rich ribosomes. Cresyl violet permanently stains a section and is suitable for both paraffin and frozen sections and subsequent RNA preparation. Besides cresyl violet, other stains like toluidine blue can be used as well.
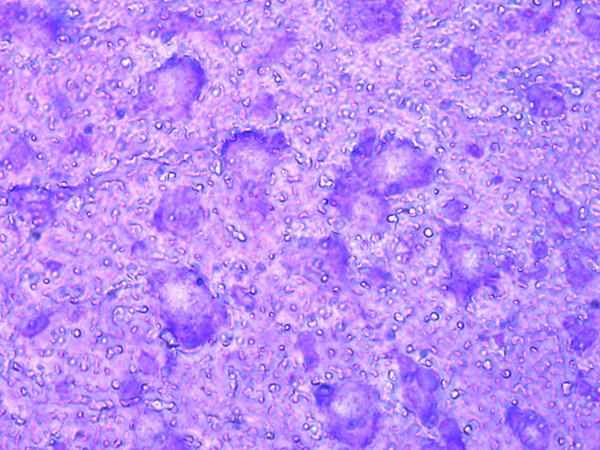
3. Visualization and ROI

Leica LMD systems can generate an automatic sample overview that helps you easily navigate to regions of interest (ROIs).
Your ROI, e.g., dopaminergic neurons, can be identified and marked automatically by the ADM software module or you can apply shapes manually.
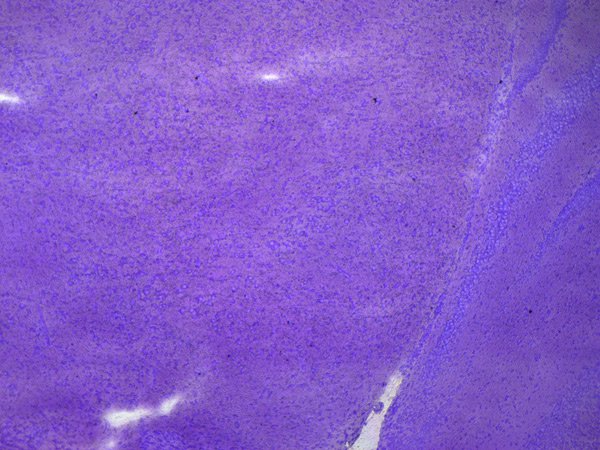
4. Laser microdissection

Next, the defined ROIs are precisely dissected by the laser under visual control and directly collected via gravity. Contact-free excision helps prevent contamination, e.g., with RNases. Also, you can cut your samples directly on the fly with the "Move and Cut" tool without any predefined shapes. The collection via gravity allows you to use standard, cost-effective consumables, such as PCR tubes or 8-strip tube caps. The collection can be done directly into a lysis buffer solution or dry air.
Leica Microsystems can provide you with the LMD system which is appropriate for your needs.
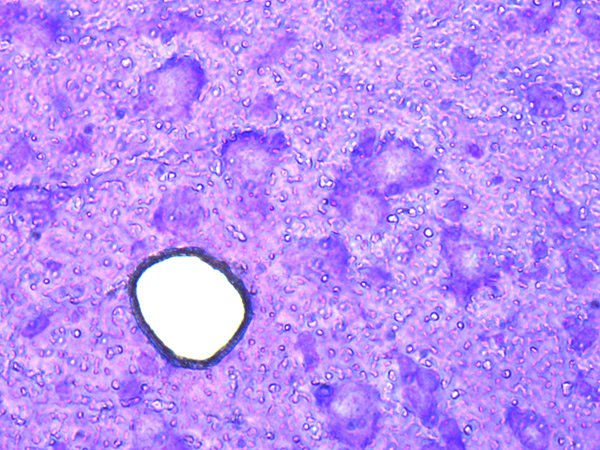
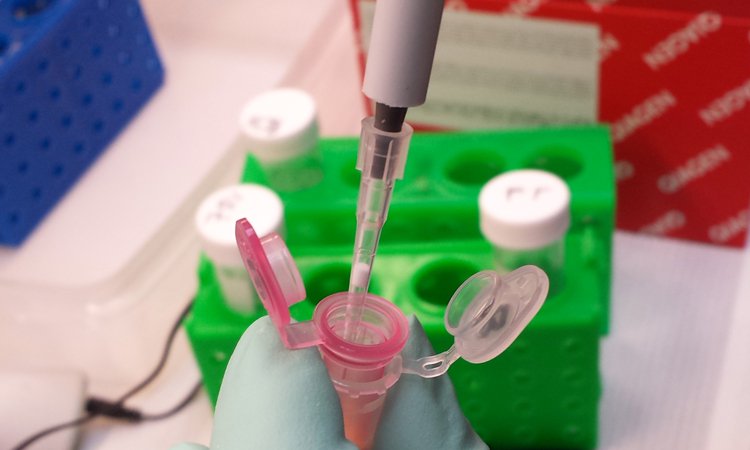
5. Extraction of RNA
Leica Microsystems recommends the high-quality QIAGEN kits (RNeasy® Micro Kit) for preparation of nucleic acids. They can be immediately used in downstream applications, such as PCR, sequencing, and quantitative and real-time PCR, or they can be stored at –20°C until needed at a later time. Please refer to www.qiagen.com for details.
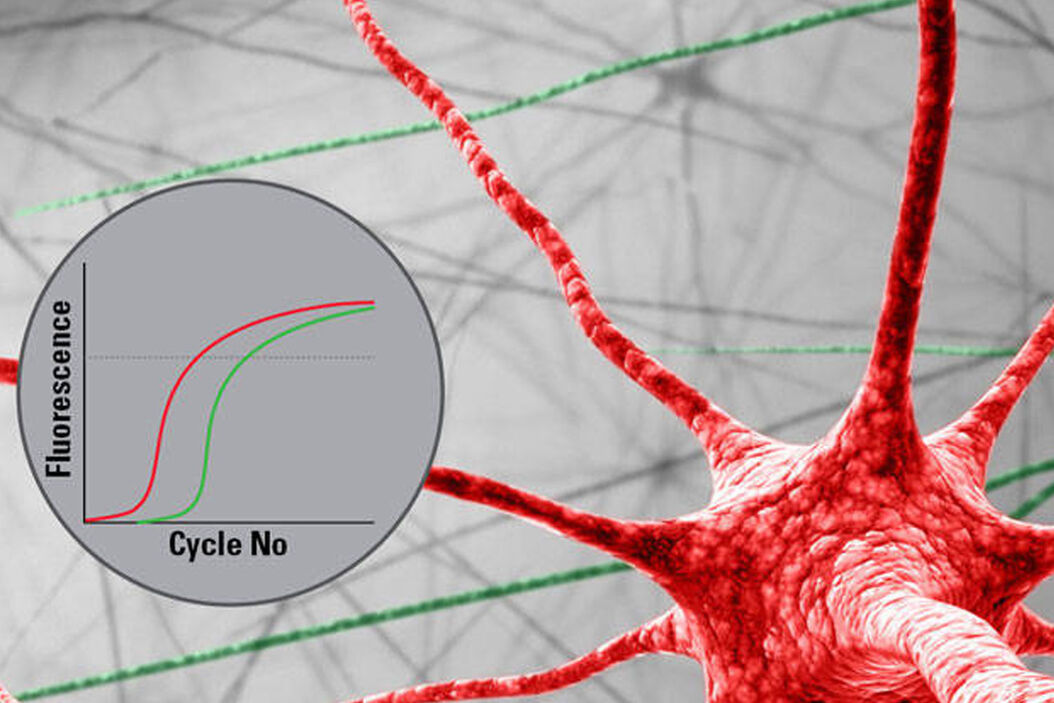
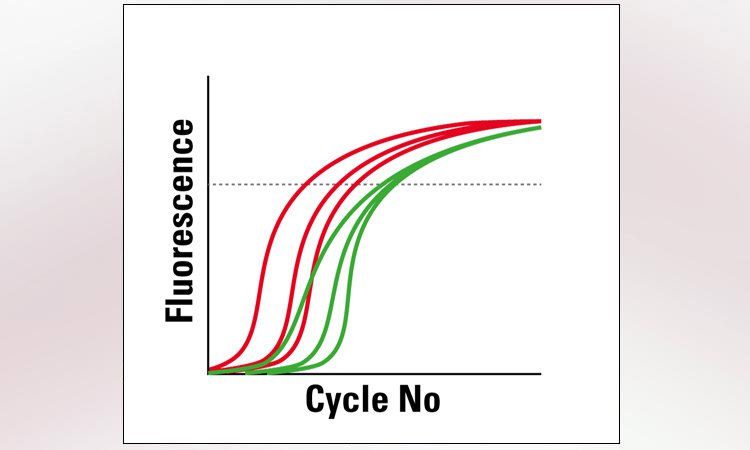
6. RNA analysis
RNA analysis is classically done via qPCR. Special LMD protocols ensure that the RNA quality remains untouched. Technologies like microarrays or next generation sequencing (NGS) are applicable as well. Thanks to the LMD cutting precision, you will get results only for the cells you are interested in. This capability enables you to compare gene expression results of different tissue areas reliably.
Typical fields of research
- Alzheimer's disease
- Developmental biology
- Aging
- Mutation analysis of tumor tissue
- Other diseases
References
- Duda, J., Fauler, M., Gründemann, J., Liss, B. Cell-Specific RNA Quantification in Human SN DA Neurons from Heterogeneous Post-mortem Midbrain Samples by UV-Laser Microdissection and RT-qPCR, In: Murray, G. (eds) Laser Capture Microdissection. Methods in Molecular Biology, vol. 1723, (Humana Press, New York, NY, 2018), pp. 335-360, DOI: 10.1007/978-1-4939-7558-7_19.
- H.-J. Lee, M.P. Jedrychowski, A. Vinayagam, S.P. Gygi, L.C. Cantley, M.W. Kirschner, Proteomic and Metabolomic Characterization of a Mammalian Cellular Transition from Quiescence to Proliferation, Cell Reports (2017) vol. 20, pp. 721–736, DOI: 10.1016/j.celrep.2017.06.074.
- H. Green, X. Zhang, K. Tiklova, N. Volakakis, L. Brodin, L. Berg, P. Greengard, T. Perlmann, P. Svenningsson, Alterations of p11 in brain tissue and peripheral blood leukocytes in Parkinson’s disease, PNAS (2017) vol. 114, iss. 10, pp. 2735-2740, DOI: 10.1073/pnas.1621218114.
- N.J. Ortner, G. Bock, A. Dougalis, M. Kharitonova, J. Duda, S. Hess, P. Tuluc, T. Pomberger, N. Stefanova, F. Pitterl, T. Ciossek, H. Oberacher, H.J. Draheim, P. Kloppenburg, B. Liss, J. Striessnig, Lower Affinity of Isradipine for L-Type Ca2+ Channels during Substantia Nigra Dopamine Neuron-Like Activity: Implications for Neuroprotection in Parkinson's Disease, Journal of Neuroscience (2017) vol. 37, iss. 28, pp. 6761-6777, DOI: 10.1523/JNEUROSCI.2946-16.2017.